In this new series of blog posts, I will talk about the architecture of the SPMF data mining library, and in particular, I will talk about the AlgorithmManager, which is a key component of SPMF, which manages all the algorithms that are provided in SPMF. I will talk about the key idea behind this module and why it is designed the way it is.
But first, let’s have a look at the overall architecture of SPMF. A picture of the architecture is given below.
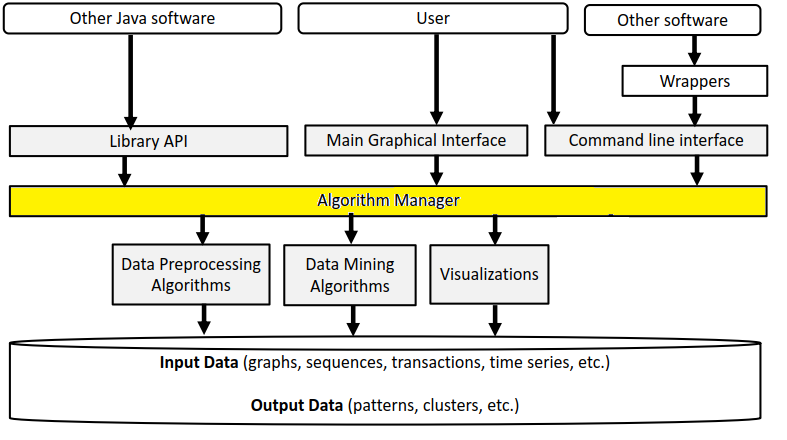
Basically, SPMF is a library of algorithms, and there are three types of algorithms: (1) algorithms for preprocessing data, (2) data mining algorithms, and (3) algorithms to visualize data or the output of algorithms, as shown by those three boxes:
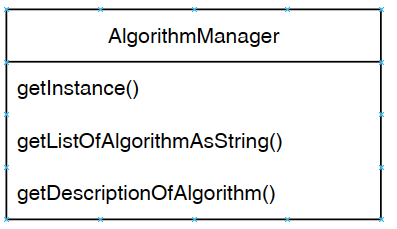
The Algorithm Manager is a key module from SPMF that manages the list of all available algorithms offered in SPMF. In particular, it provides the list of all algorithms to the user interface and command line interface of SPMF.
The three main methods (functions) of the Algorithm Manager are illustrated below:
To access the algorithm manager from the Java code, we must write. AlgorithmManager.getInstance() to obtain the instance of the Algorithm Manager. Then, we can call two key methods (functions) of the algorithm manager, which are:
- getListOfAlgorithmsAsString(): returns the list of all algorithms that are offered in SPMF (as a list of strings),
- getDescriptionOfAlgorithm(): returns the description of an algorithm that has a given name, which allows to know more about the algorithm and also to run the algorithm.
I will next show you some examples about how to use these two functions, while providing more explanations.
Example 1: Obtaining the list of all algorithms offered in SPMF
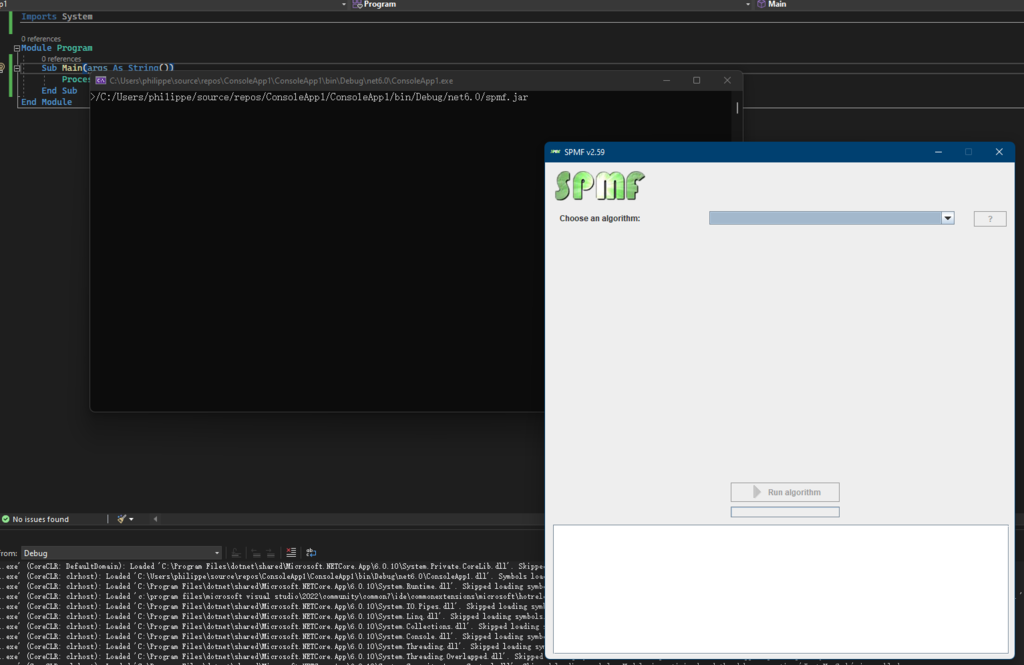
First, let me show you an example of how to use the AlgorithmManager to obtain the list of algorithms offered in SPMF. Here I wrote a small Java program:
import java.util.List;
import ca.pfv.spmf.algorithmmanager;
public class Example1{
public static void main(String[] args) throws Exception {
List<String> list = AlgorithmManager.getInstance().getListOfAlgorithmsAsString(true, true, true);
for(String name : list) {
System.out.println(name);
}
}
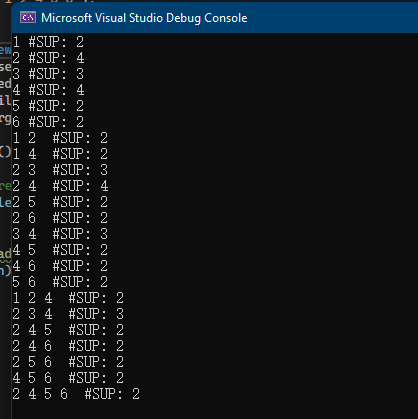
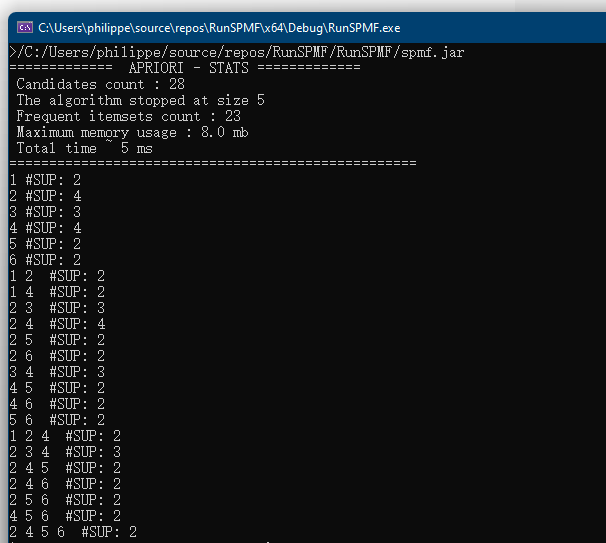
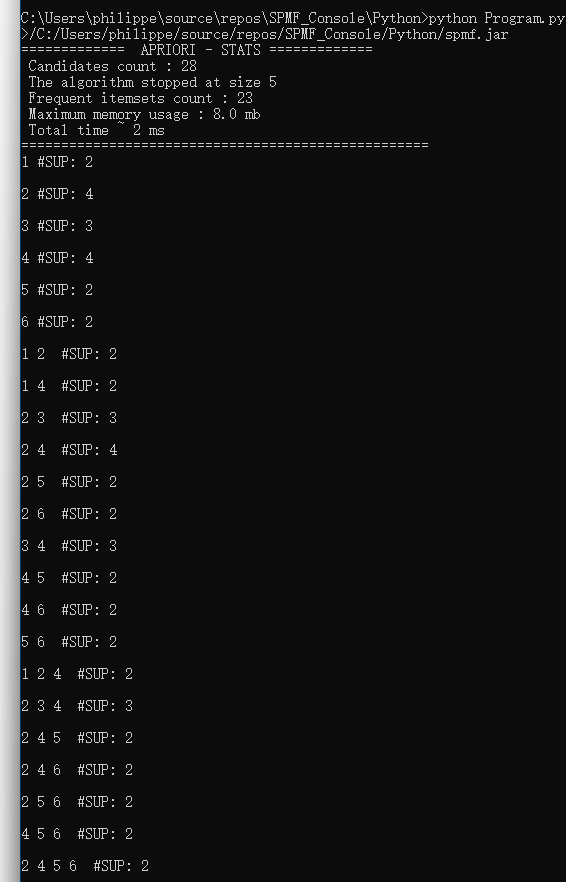
}By running this program, the list of available algorithms from SPMF will be printed in the console like this:
If you look carefully at this output, you will notice that there are two types of elements in that list: (1) names of algorithms (e.g. “Apriori_association_rules”), and (2) names of categories of algorithms (starting with ” — “). For example, in the category ” — CLUSTERING — “, there are several clustering algorithms such as “BisectingKMeans”, “DBScan”, “Hierarchical_clustering” etc. The algorithms are classified into categories to make it easier for users to look for algorithms.
Another thing that you may notice in the above example, is that the method “getListOfAlgorithmsAsString()” has three Boolean parameters:
getListOfAlgorithmsAsString(true, true, true);
Why? Those Boolean parameters are filters. Setting them to true indicate that we want to list all algorithms from the three types of algorithms (the (1) preprocessing algorithms, (2) the data mining algorithms, and (3) the algorithms for visualizations). If we want to see only the data mining algorithms, we would change as follow:
getListOfAlgorithmsAsString(false, true, false);
Example 2: Obtaining information about a specific algorithm
Now, let me show you a second example, where I will explain how to obtain information about a specific algorithms from SPMF. Here is a simple Java program that calls the AlgorithmManager to get information about the “RuleGrowth” algorithm and print the information to the console:
import java.util.Arrays;
import ca.pfv.spmf.algorithmmanager;
public class Example2{
public static void main(String[] args) throws Exception {
// / Initialize the algorithm manager
AlgorithmManager algoManager = AlgorithmManager.getInstance();
DescriptionOfAlgorithm descriptionOfAlgorithm = algoManager.getDescriptionOfAlgorithm("RuleGrowth");
System.out.println("Name : " + descriptionOfAlgorithm.getName());
System.out.println("Category : " + descriptionOfAlgorithm.getAlgorithmCategory());
System.out.println("Types of input file : " + Arrays.toString(descriptionOfAlgorithm.getInputFileTypes()));
System.out.println("Types of output file : " + Arrays.toString(descriptionOfAlgorithm.getOutputFileTypes()));
System.out.println("Types of parameters : " + Arrays.toString(descriptionOfAlgorithm.getParametersDescription()));
System.out.println("Implementation author : " + descriptionOfAlgorithm.getImplementationAuthorNames());
System.out.println("URL: : " + descriptionOfAlgorithm.getURLOfDocumentation());
}
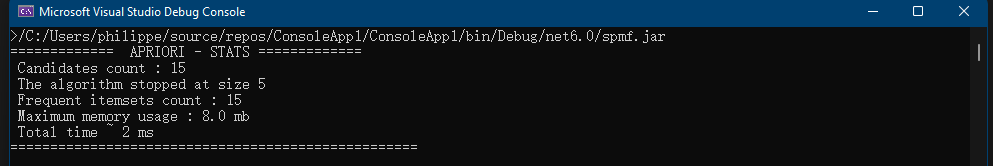
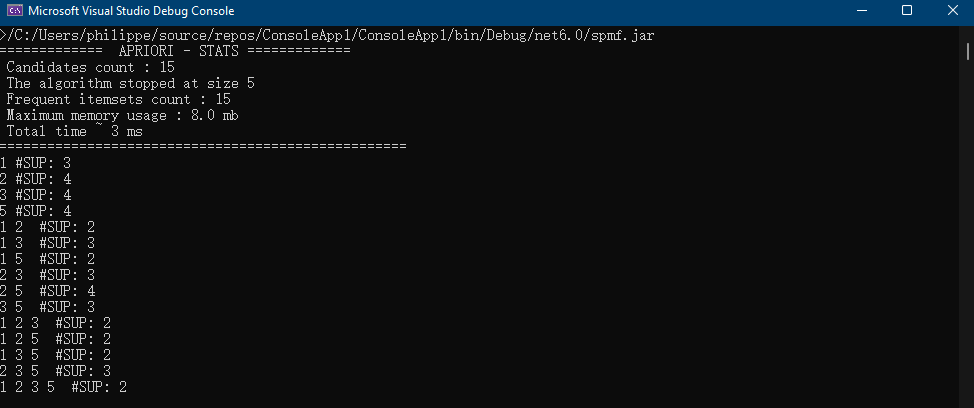
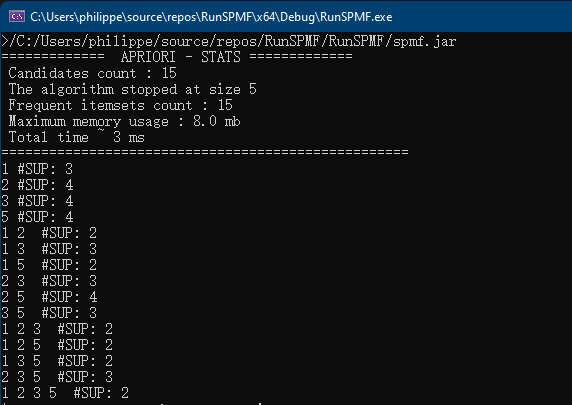
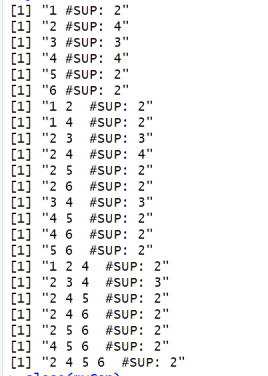
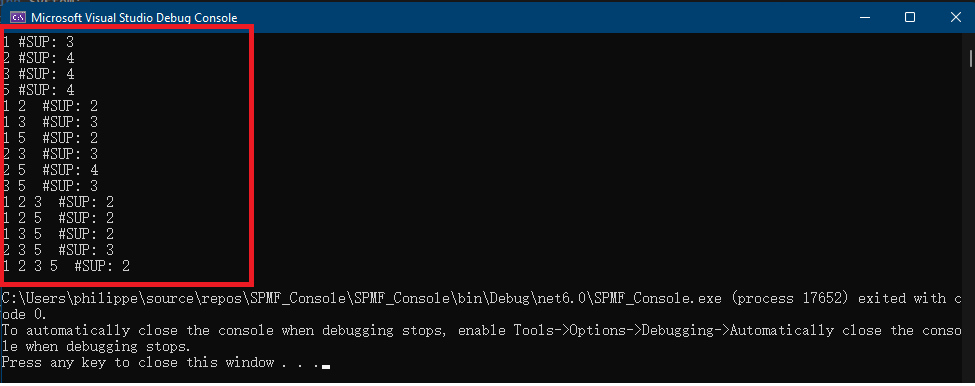
}The result of running this code is that information about the RuleGrowth algorithm is printed in the console:
Name : RuleGrowth Category : SEQUENTIAL RULE MINING Types of input file : [Database of instances, Sequence database, Simple sequence database] Types of output file : [Patterns, Sequential rules, Frequent sequential rules] Types of parameters : [[Minsup (%), (e.g. 0.5 or 50%), class java.lang.Double, isOptional = false ], [Minconf (%), (e.g. 0.6 or 60%), class java.lang.Double, isOptional = false ], [Max antecedent size, (e.g. 1 items), class java.lang.Integer, isOptional = true ], [Max consequent size, (e.g. 2 items), class java.lang.Integer, isOptional = true ]] Implementation author : Philippe Fournier-Viger URL: : http://www.philippe-fournier-viger.com/spmf/RuleGrowth.php
This output indicates the name of the algorithm, the category that it belongs to, its type of input file and output file, the type of parameters that it takes, who is the implementation author and an URL to the documentation of SPMF for that algorithm.
Now lets me explain in more details about how it works.
When we call the method algoManager.getDescriptionOfAlgorithm(“RuleGrowth“), the Algorithm Manager returns an object of type DescriptionOfAlgorithm. The class DescriptionOfAlgorithm is an abstract class designed to store information about any algorithm. Each algorithm in SPMF must have a subclass of DescriptionOfAlgorithm that provide information about the algorithm.
For example, for the RuleGrowth algorithm, there is a class DescriptionAlgoRuleGrowth that is a subclass of DescriptionOfAlgorithm, which provides information about the RuleGrowth algorithm. If you are curious, here is the code of that class:
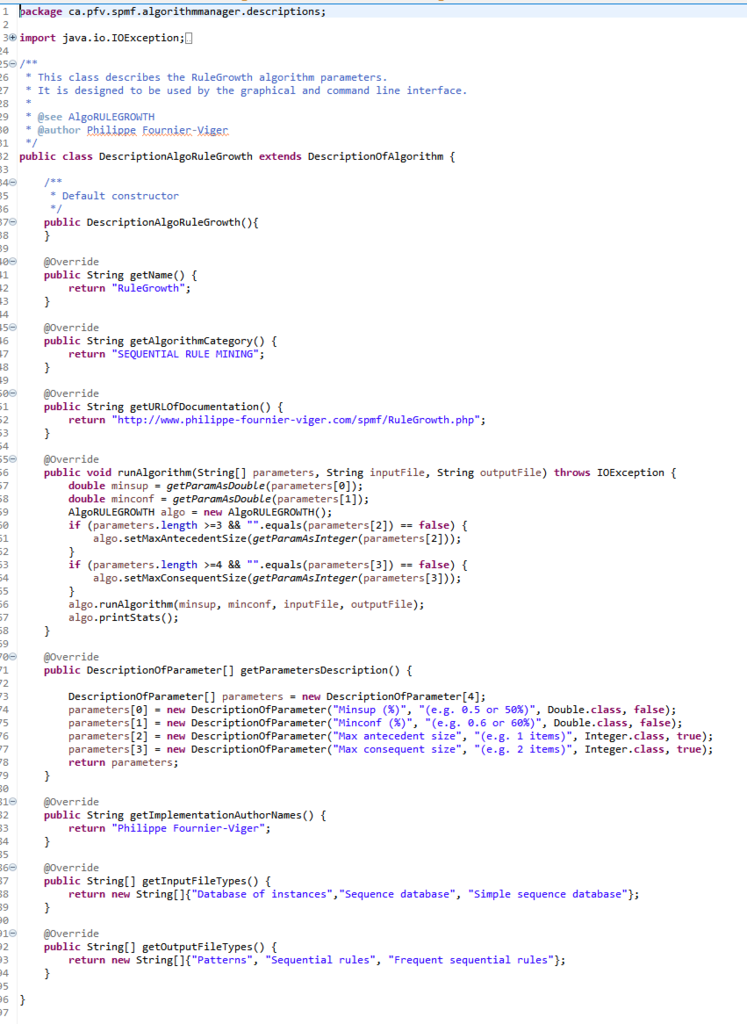
Each subclass of DescriptionOfAlgorithm must implement a set of methods to provide information about the algorithm. Those methods are:
- getName(): return the name of the algorithm (e.g. RuleGrowth)
- getAlgorithmCategory(): return the category of the algorithm (e.g. SEQUENTIAL RULE MINING)
- getURLOfDocumentation(): return an URL to a webpage describing this algorithm
- runAlgorithm(): this method is used to call this algorithm (apply it)
- getParameterDescription(): obtain information about all the parameters of the algorithm. This is provided as a list of object of type DescriptionOfParameter. Basically, for each parameter, we have a name, an example, the type of parameter (e.g. Double) and a Boolean indicating if the parameter is optional (e.g. true) or not.
- getImplementationAuthorNames(): returns the name(s) of who implemented the algorithm
- getInputFileTypes(): return the types of input files that this algorithm take as input. It is a list of String from the most general type to the most specific.
- getOutputFileTypes():return the types of output files that this algorithm take as input. It is a list of String from the most general type to the most specific.
To summarize, here is an illustration of the relationship between the Algorithm Manager and the classes DescriptionOfAlgorithm and DescriptionOfParameter:
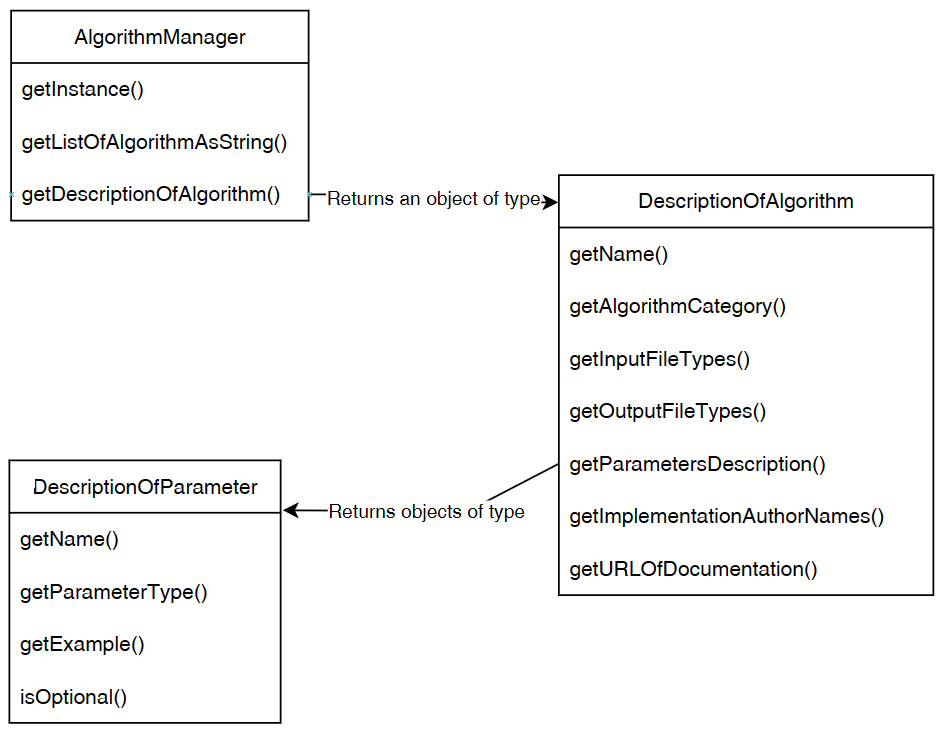
Example 3: Running an algorithm
Now let me show you how to use the algorithm manager to run an algorithm from SPMF. Lets look at the following example:
import ca.pfv.spmf.algorithmmanager;
public class Example3{
public static void main(String[] args) throws Exception {
AlgorithmManager algoManager = AlgorithmManager.getInstance();
DescriptionOfAlgorithm descriptionOfAlgorithm = algoManager.getDescriptionOfAlgorithm("PrefixSpan");
String[] parameters = new String[]{"0.4","50","true"};
String inputFile = "contextPrefixSpan.txt";
String outputFile = "./output.txt";
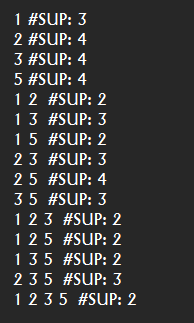
descriptionOfAlgorithm.runAlgorithm(parameters, inputFile, outputFile);
}
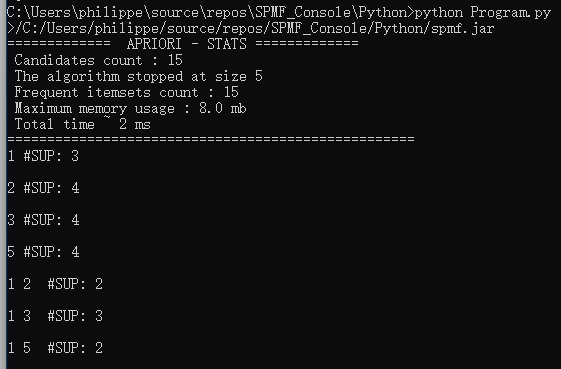
}This Java program calls the function getDescriptionOfAlgorithm to first obtain the description of the PrefixSpan algorithm. Then, the program call the method runAlgorithm() of that description to execute the algorithm on a file called “contextPrefixSpan.txt” and save the result in a file “output.txt”. Note that to run this example, it is necessary that the file “contextPrefixSpan.txt” is located in the right location on your computer or that you give the full path to the file.
Any algorithms from SPMF can be called in a similar way through the AlgorithmManager.
More about the Algorithm Manager
Now that you know more about the AlgorithmManager and its purpose, let me explain a bit more about the internal design of the AlgorithmManager.
When I implemented this module, I wanted to avoid having a hard coded list of algorithms in the code to make the software easier to maintain. Thus, I have decided that each algorithm in SPMF would instead have a class that describes it, which is a subclass of DescriptionOfAlgorithm. For example, the RuleGrowth algorithm has a class DescriptionAlgoRuleGrowth to describe the RuleGrowth algorithm.
Now, internally, to avoid hard coding the list of all algorithms, the AlgorithmManager scans the package “ca.pfv.spmf.algorithmmanager.descriptions;” to automatically find all the subclasses of DescriptionOfAlgorithm. This allows to automatically find all algorithms that are available in SPMF and make the list of them. The AlgorithmManager can then give this list to the user interface of SPMF, etc.
Thus, if we want to add a new algorithm to SPMF, we just need to create a new subclass of DescriptionOfAlgorithm and put it in the package “ca.pfv.spmf.algorithmmanager.descriptions;” and the Algorithm Manager will automatically detect it, which is very convenient.
If you are curious, this detection is done by the following code in the AlgorithmManager class:

which calls this function:
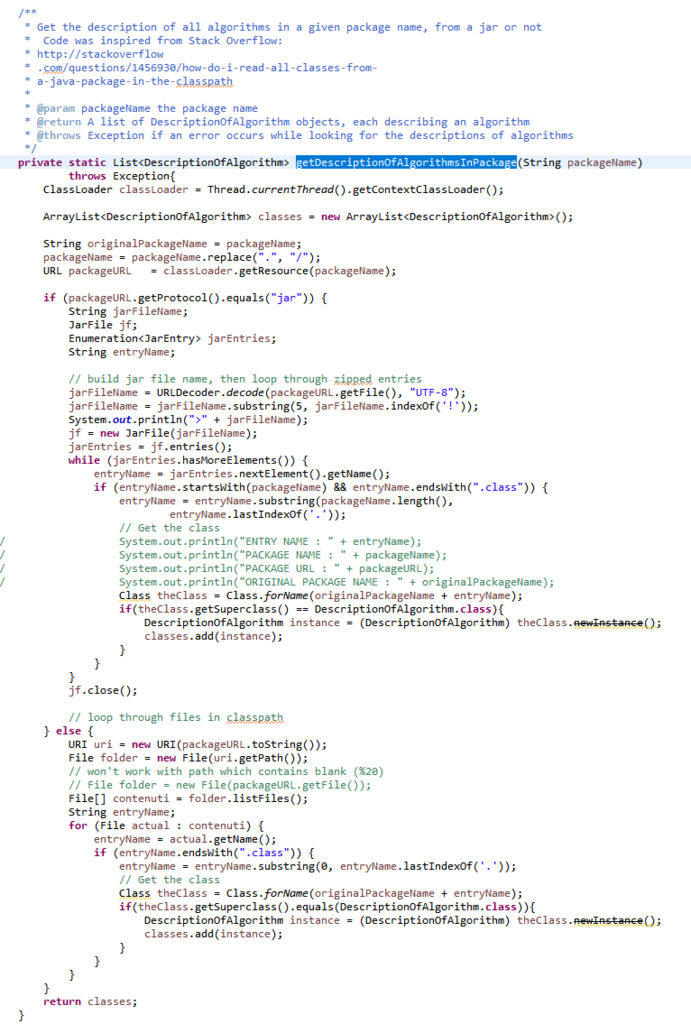
The above function is somewhat complex because it has to work both when SPMF is called as JAR file or when it is called from the source code. I will not explain this code in more details.
Conclusion
In this blog post, I have given an overview of a key module in SPMF, which is the AlgorithmManager. In upcoming blog posts, I will explain other interesting aspects of the architecture of SPMF. Hope that this has been interesting.
==
Philippe Fournier-Viger is a full professor and the founder of the open-source data mining software SPMF, offering more than 250data mining algorithms.