In this blog post, I will talk about the future of SPMF. I do this once in a while. Today, I will discuss on the performance of SPMF.
Performance
A main focus of SPMF is and has always been efficiency in memory and time. This is something that is different from many other data mining software, which do not focus much on efficiency. For example, a few years ago, I did a performance comparison of some popular algorithms implemented in SPMF with those implemented in two other software: Weka and Coron, also coded in Java, and the difference in terms of runtime for the same algorithms was huge (the comparison can be found on that page). For example, here is the difference between the FP-Growth algorithm implemented by SPMF and by Weka on some benchmark datasets called “Chess” and “Mushrooms”:
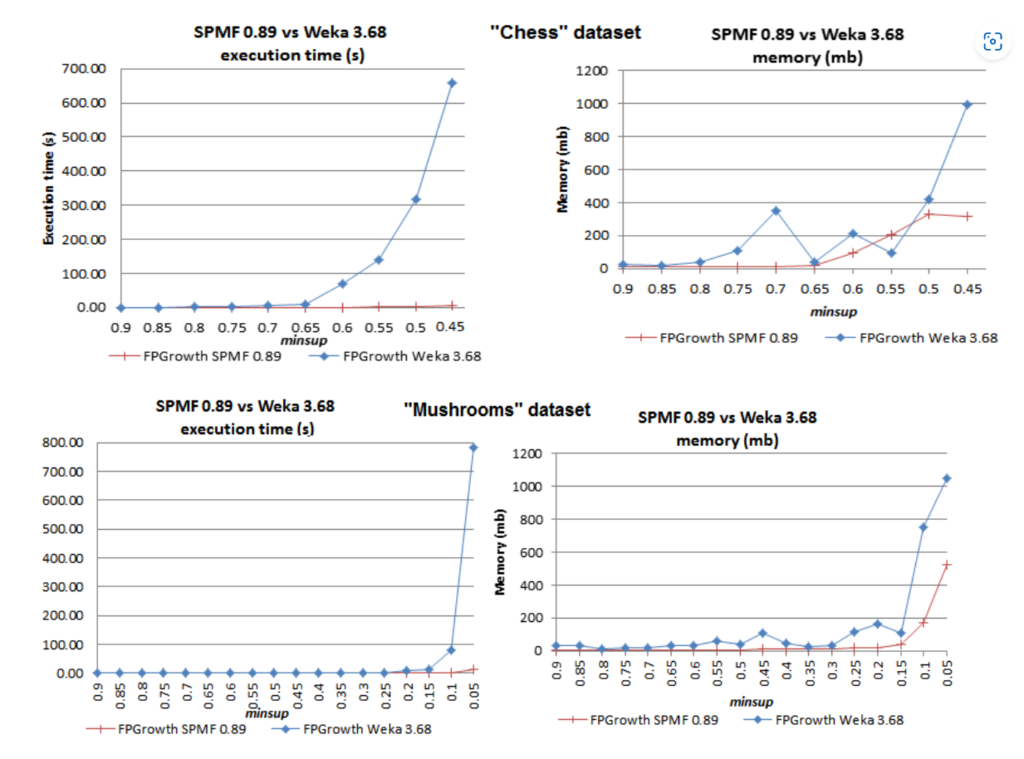
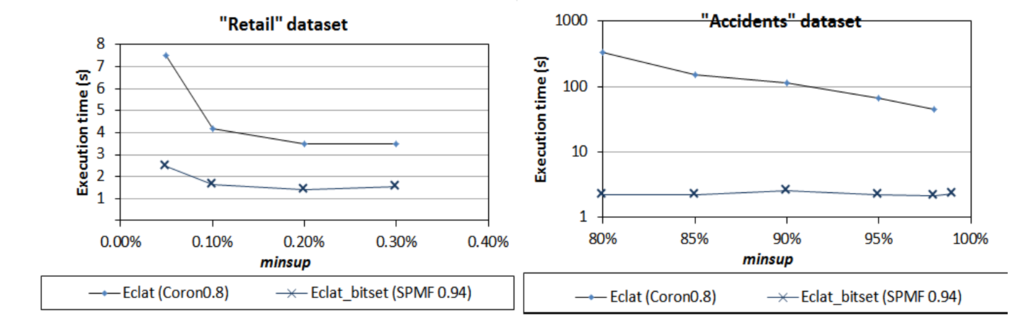
As we can see, SPMF’s version of FP-Growth can be more than 100 times faster than Weka. This shows that, at that time at least, the version of Weka was poorly optimized. And for Coron, we found the gap to be smaller, but there was sometimes quite a big difference as well in some cases. For example, here is a performance comparison for the Eclat algorithm:
Some other people have also done performance comparison between SPMF and other data mining software. For example, I have found a paper “dbscan: Fast Density-based Clustering with R” where the performance of SPMF’s version of the DBScan and Optics algorithms for clustering was compared with many other libraries such as scikit, weka, elki, and pycluster. The results from that paper:
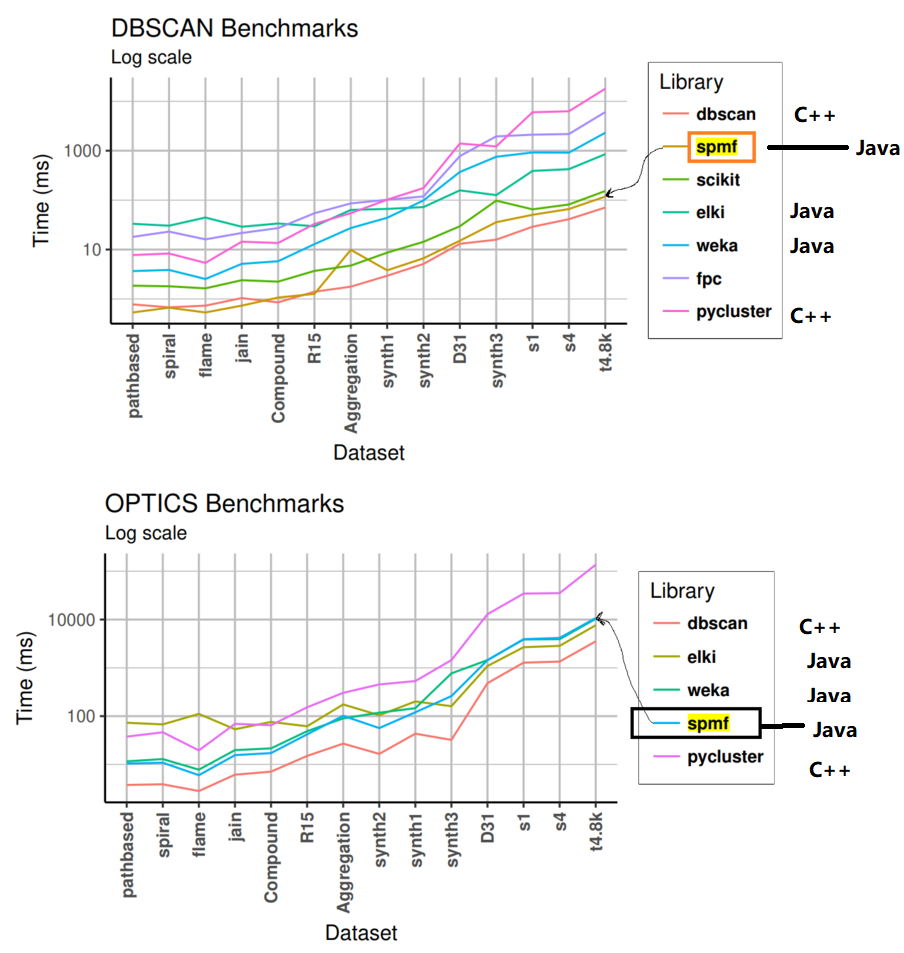
As can be seen in these figures, SPMF is very fast. It is generally the second fastest and generally very close to the first. This is very good considering that clustering is not the main focus of SPMF and that SPMF is implemented in Java, while the first one is implemented in C++. Thus, despite being implemented in Java, SPMF can pretty much match the speed of the C++ version. In these results, we can also see that SPMF is faster than Weka (again) and also that SPMF is generally faster than Eki, although that latter is also implemented in Java and specialized in clustering.
Why SPMF is efficient?
For SPMF, from the beginning, I have paid much attention to optimizing the code as much as possible for all aspects. This has been done by making careful decisions when implementing algorithms, and to use appropriate techniques. Generally, most algorithms are well optimized, and especially the most popular ones. But also a major reason is that SPMF is very lightweight as it does not depend on external libraries. Besides, the structure of SPMF has always been kept simple to ensure high performance. For example, it would have been possible to use a complex software architecture with lot of classes, interfaces, and many libraries that provide many functions that are not needed, but that would have impacted the performance.
For the future? More efficiency!
Now, let me talk about the future of SPMF. A part of it will be yet more efficiency. Currently, I am working on a major improvement of SPMF that will increase even more the performance. How? One way is that I have recently developed a new set of data structures that are highly optimized for SPMF to replace the default ArrayList, HashMap and HashSet classes offered by Java. Moreover, some additional specialized data structures will be added such as cache structures, efficient bitsets, etc, with custom iterators, comparators, etc. for higher speed and lower memory usage.
I will not share the details today as I am still working on polishing the code and testing as I do not want to release unstable code. Moreover, I need to replace the data structures from existing algorithms in SPMF by the new one, which may take several weeks as there are hundreds of algorithms. But this should be released in the next major version of SPMF, which should be called SPMF 3.0.
And there will be a considerable performance gain for some algorithms, especially in terms of memory usage. For example, just by replacing the Java HashMap and ArrayList data structures by the new optimized versions of HashMap and ArrayList from SPMF, I have quickly made a modified version of the ULB-Miner algorithm yesterday and the memory usage was reduced by 73%. For example, here is the result on the Chess dataset for minutil = 500000:
ULB-MINER BEFORE, CHESS, minutil = 500000
Total time ~ 11471 ms
Memory ~ 99.1240234375 MB
High-utility itemsets count : 24979
ULB-MINER AFTER, CHESS, minutil = 500000
Total time ~ 21449 ms
Memory ~ 27.316810607910156 MB (73 % less memory usage)
High-utility itemsets count : 24979
Here we can see about a 73% reduction of memory usage, which is very significant. And I actually did this in about 30 min. I could improved it further and do more improvements. I think that this type of optimization can make a significant performance difference for several algorithms.
If you are a researcher and you are interested in testing the new optimized data structures of SPMF before they are released (maybe in 1 or 2 months), you may leave me a message and I could share that with you, if you want to provide me some feedback to help me improve that before the release. I think from a research perspective this is also quite interesting, as it can help boost the performance of a new algorithm.
That is all for today. It was just a quick update about SPMF.
==
Philippe Fournier-Viger is a full professor and the founder of the open-source data mining software SPMF, offering more than 250data mining algorithms.