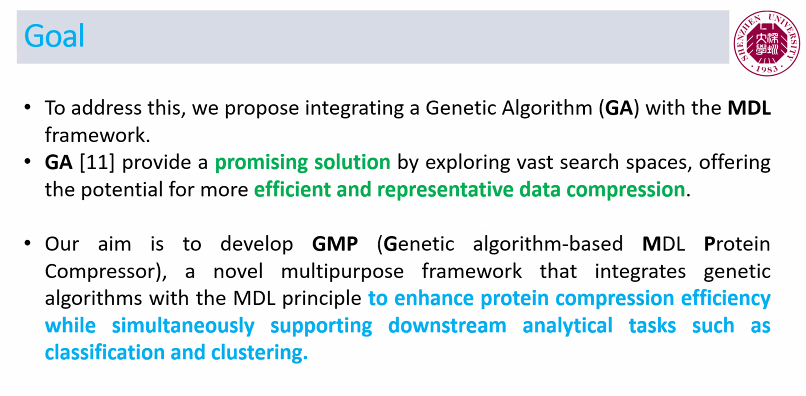
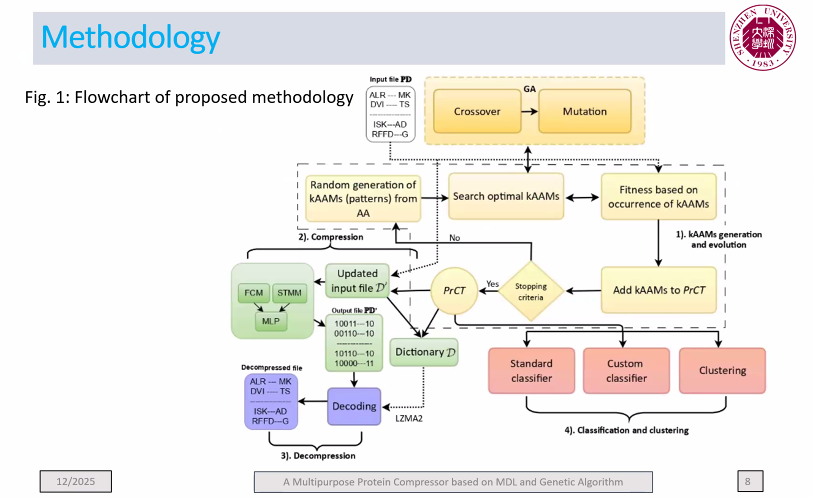
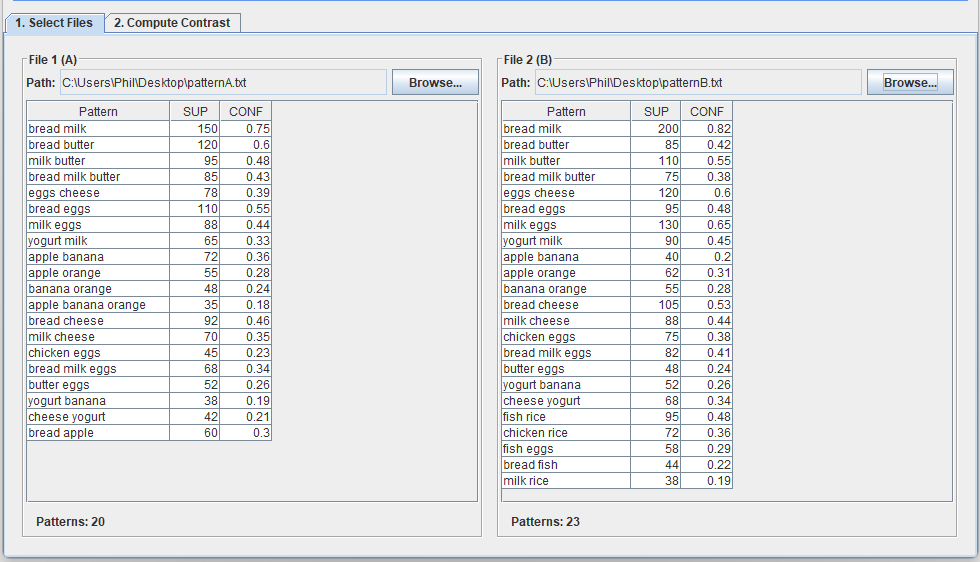
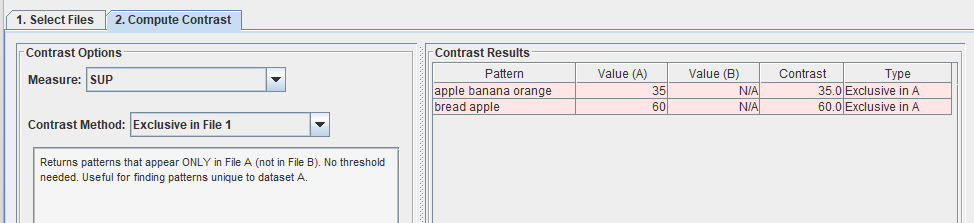
Today, I want to talk to you about something quite incredible. I recently attended a keynote speech at an international conference. The keynote speaker was presenting online and was scheduled for a 35 minute presentation. However, he had prepared 180 slides. Yes, you have read that correctly: 180 slides!
This is a ridiculous number of slides for a talk. Generally, a good rule of thumb for a presentation is to prepare one slide per minute. If we follow this rule, a 35 minute presentation should have 35 slides. So, as I would expect in such case, the talk went overtime…
After 1 hour (already 25, the speaker was at slide 140, while going at a speed of 2.3 slides per minute!
Then, a few minutes later, the organizer intervened to stop the talk. And the speaker said sorry for going overtime…. But seriously, how can you be sorry when you prepare 180 slides.
I have seen some speaker exceed the time allowed at several conferences. However, usually, it is by 5 or 10 minutes, rarely by over half an hour. And if he had not been stopped, at this speed, it would have taken almost 1 hour and half to finish the presentation.
Personally, I do not understand this. If you are invited to give a talk at a conference, I think it is important to follow the schedule, so as to show respect to the organizers, to the audience and to the speakers that will talk after you. Because of that speaker, other speakers had to shorten their presentations, and the coffee break had to be shortened, as well.
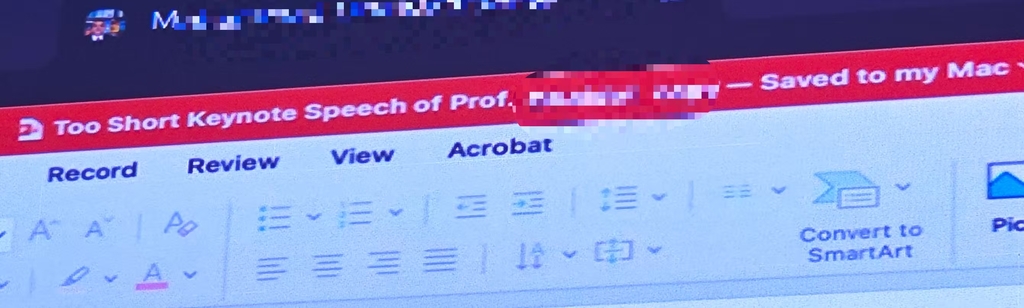
What’s more, it seems that the professor was well aware that he had too many slides as the title of his presentation was ironically titled “The too short keynote speech of Prof (…. ) “, as you can see in the picture below:
I will not say the name of the speaker. But he is a senior Jordanian professor with over 20,000 citations.
So my goal by telling this story is just to remind everyone that it is important to follow the time constraints at conferences. It shows respect and professionalism. That is all I wanted to say for today! If you want to share your opinion, please leave a comment below.