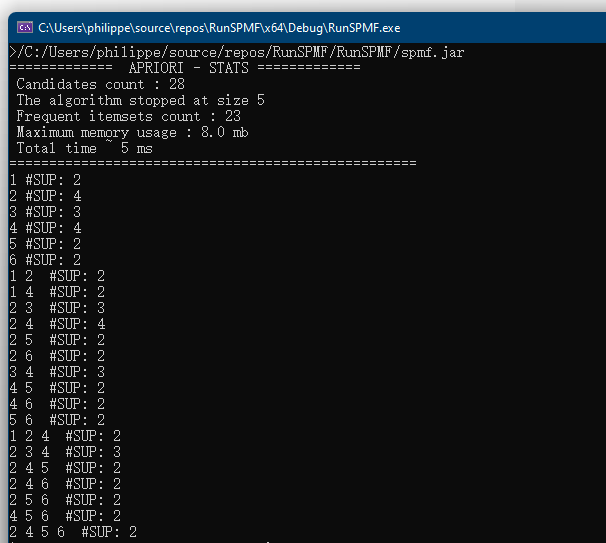
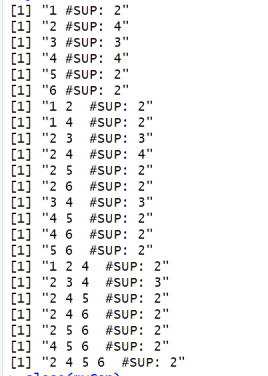
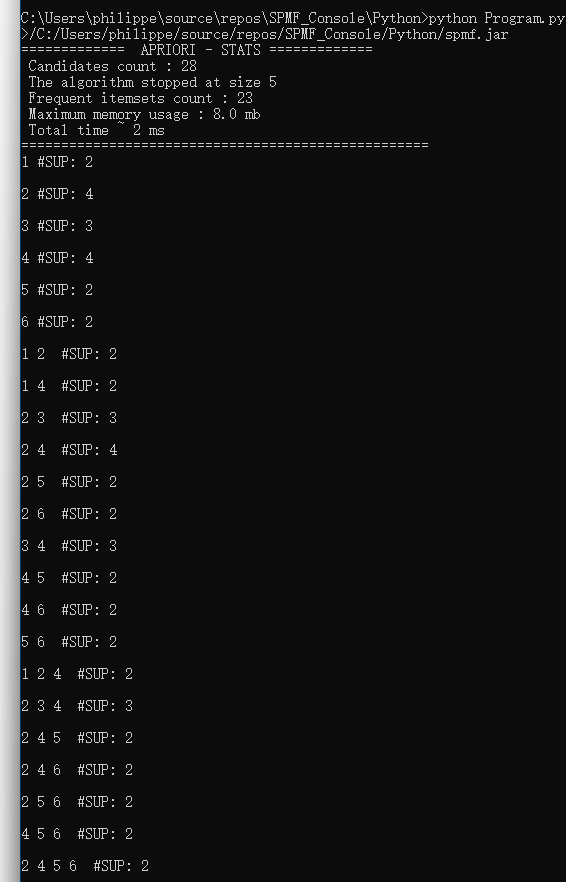
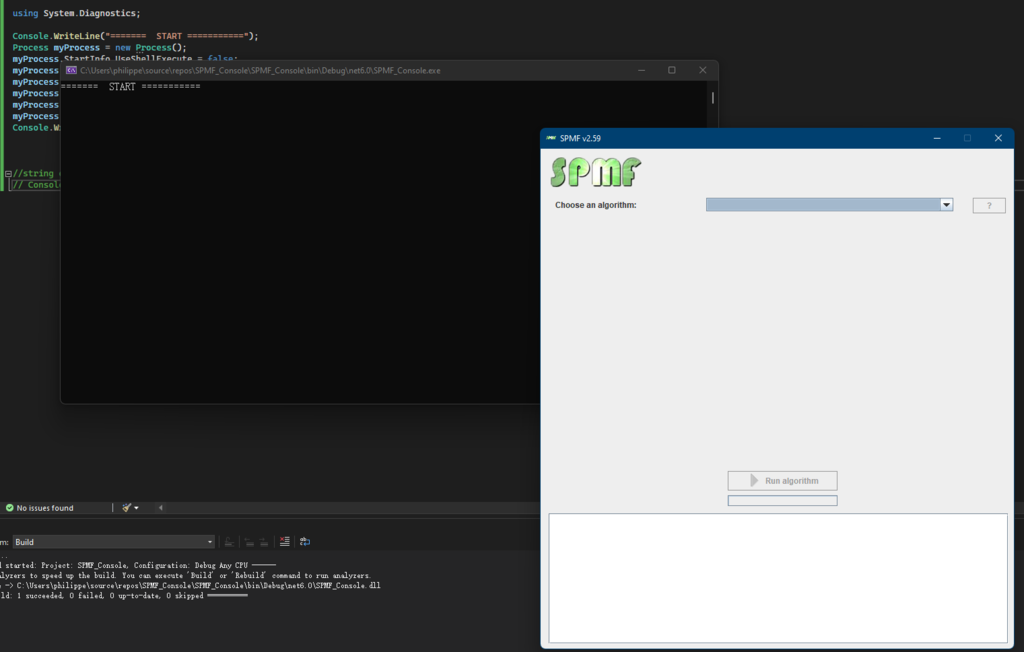
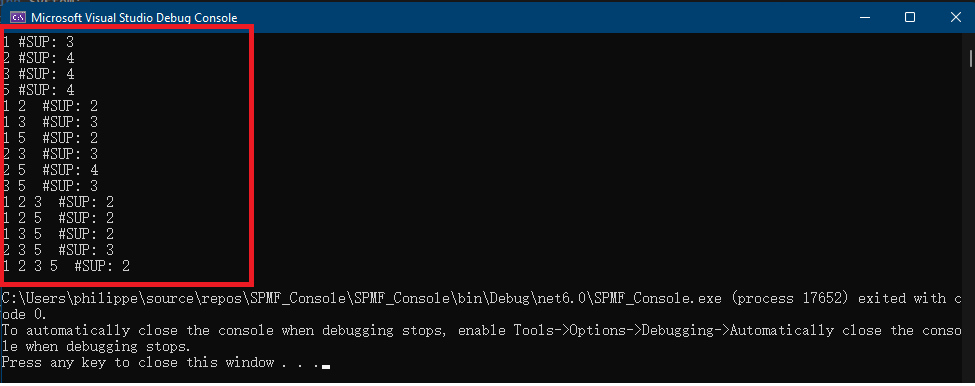
In this blog post, I will continue explaining the architecture of the SPMF data mining library. In the previous post, I have introduced a key component of SPMF called the Algorithm Manager, which manages all the algorithms offered in SPMF.
Today, I will move on to talk about two other key components in the architecture of SPMF. In particular, I will focus on how SPMF can be run from both a graphical interface and a command line interface. How does these two interfaces can work seamlessly with the rest of the code in SPMF? The short answer is that it is thanks to two modules called the Main class and the Command Processor, that I will explain in this article. Briefly, the Main class is the entry point for running SPMF, which detects whether SPMF is started from the command line or not, and then launches the graphical interface or the command line interface. And the Command Processor is the module that take care of running a command (e.g. executing a data mining algorithm or launching a visualization). A command is either launched by the command line or graphical interface.
A brief overview of SPMF’s architecture
Before explaining this in details, let’s briefly review the overall architecture of SPMF.
SPMF is a Java software, and it is distributed as a JAR file, as most software implemented in Java:
SPMF is designed to be used in three ways:
- as a Java library that can be imported in other Java project
- as a standalone program with a simple graphical user interface
- as a standalone program that can be run from the command line
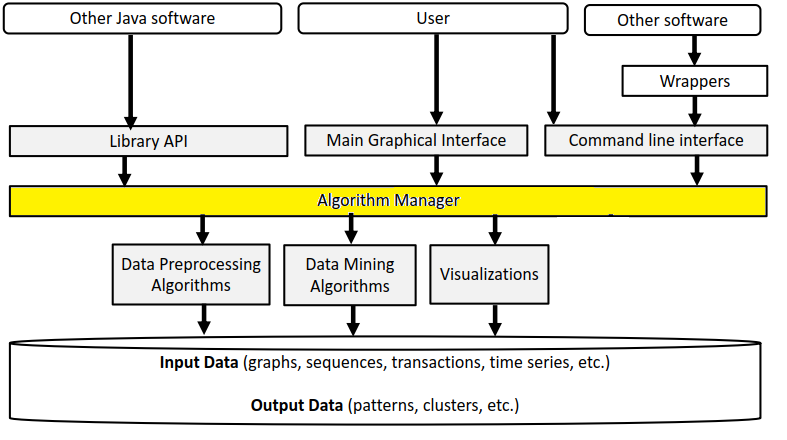
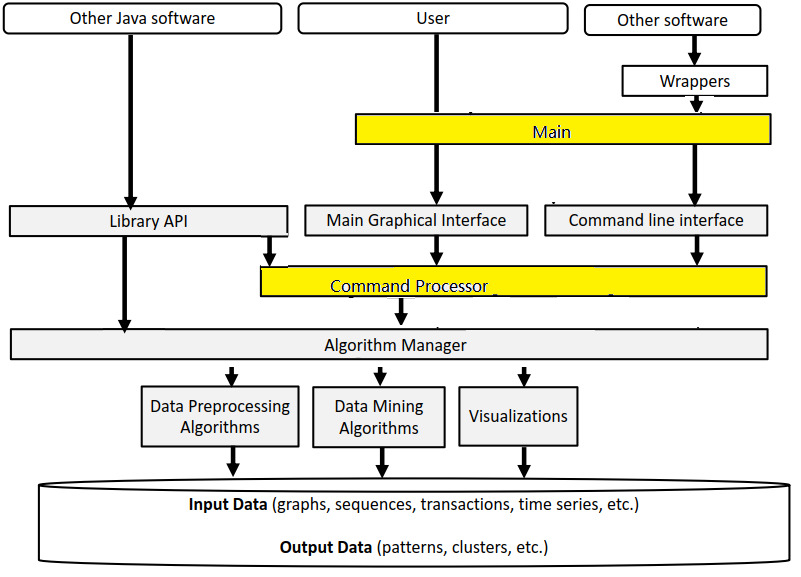
The architecture of SPMF is presented in the figure below:
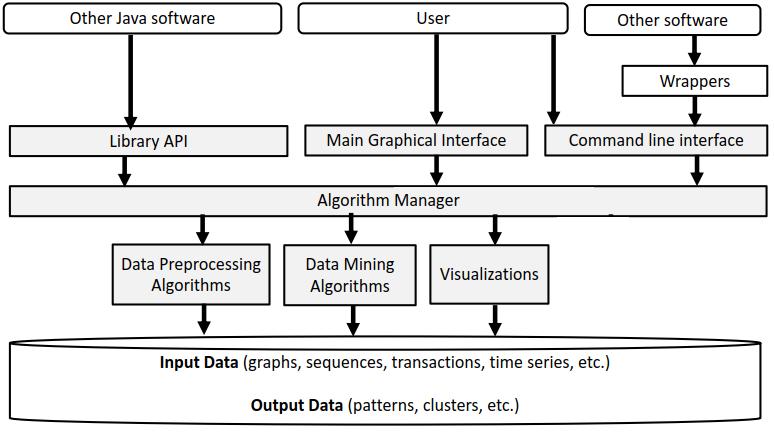
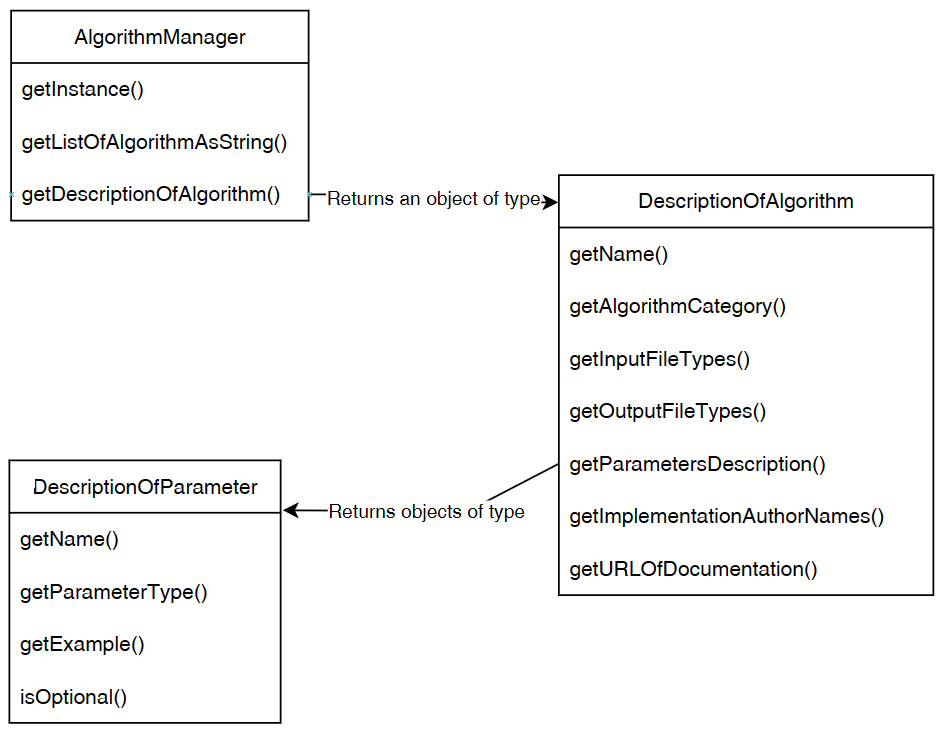
As can be seen in the top of this figure, the SPMF software can be called by other Java software or by the user using the library API, graphical interface or command line interface. Then, all these interfaces rely on a class called the Algorithm Manager to obtain information about the available algorithms and how to run them. There are three types of algorithms: (1) preprocessing algorithms and tools, (2) data mining algorithms and (3) visualizations.
The Main Class
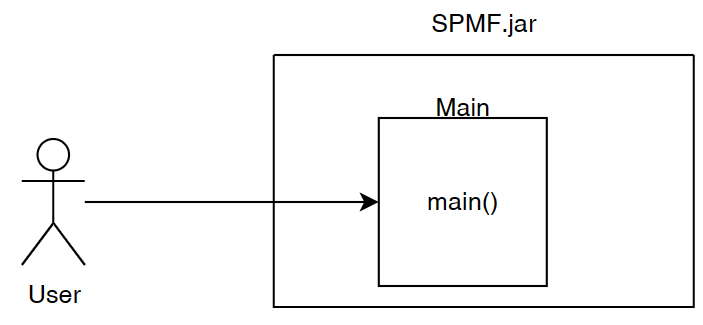
Now let’s get into details. As I said previously, the SPMF software is packaged and distributed as a JAR file. To make a Jar file that can be executed as a program, it is necessary to choose a Main class that will be the entry point for the program.The Main class play this role. It is located in the package ca.pfv.spmf.gui of SPMF.
When the user launches SPMF program from the command line or by double-clicking on the JAR file to start the graphical user interface, the method main() of the class Main is called.
Here is the code of the main() method:
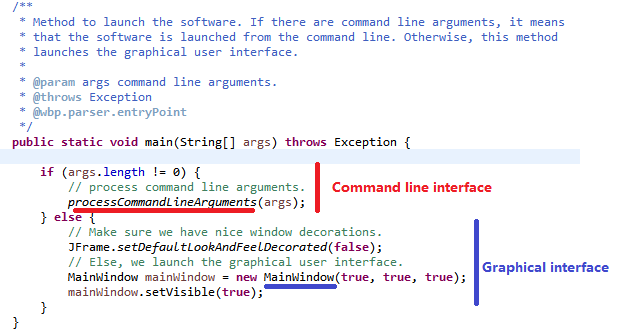
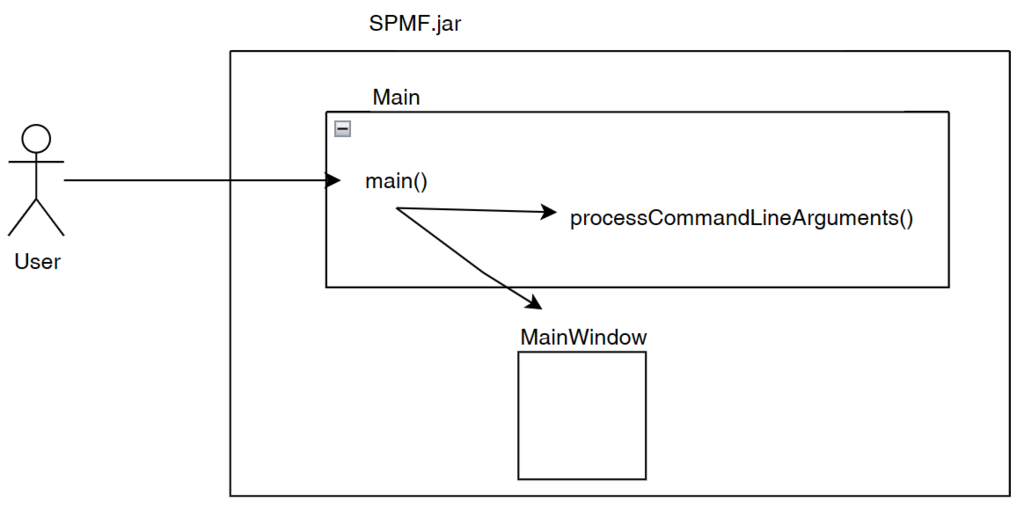
Briefly, the method checks if some arguments have been passed to the program. If there aresome arguments, it means that SPMF is executed from the command line. Thus, the method processCommandLineArguments(args) is called to execute the command that is received from the command line. Otherwise, if there is no argument, it means that the user wants to launch the graphical interface. In that case, the main window of SPMF is created which is called MainWindow in the current version of SPMF and it is displayed to the user.
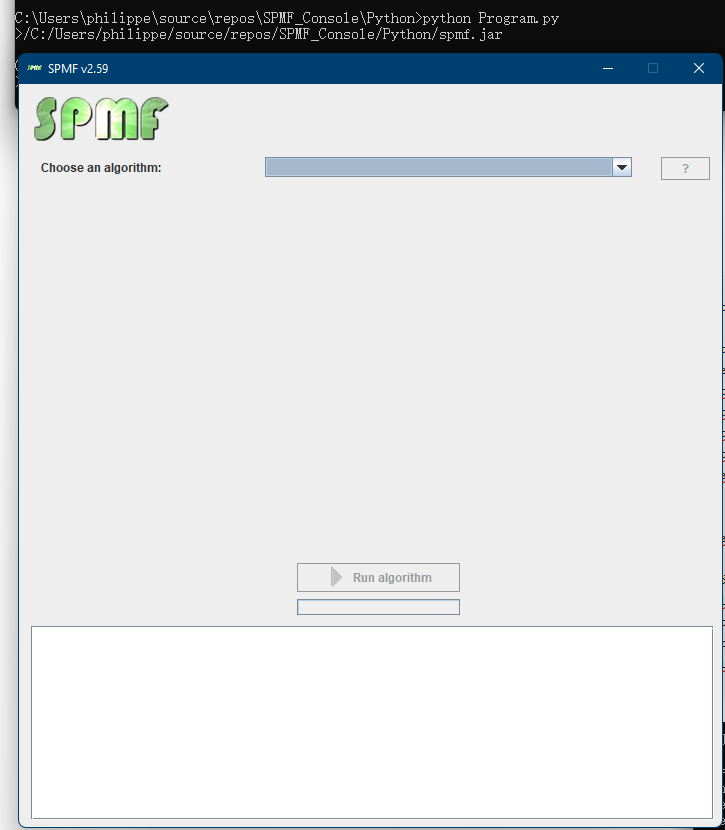
This is the MainWindow:
The Command Processor
Another important module in SPMF is the Command Processor. It is a class that is shared by both the command line interface and graphical user interface. The Command Processor is used to run algorithms that the user has selected either from the command line or graphical user interface.
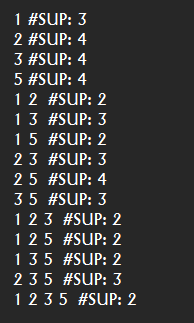
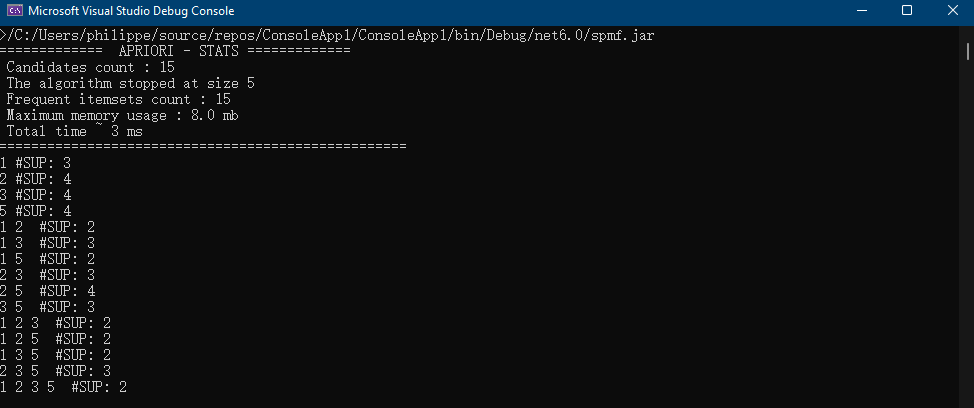
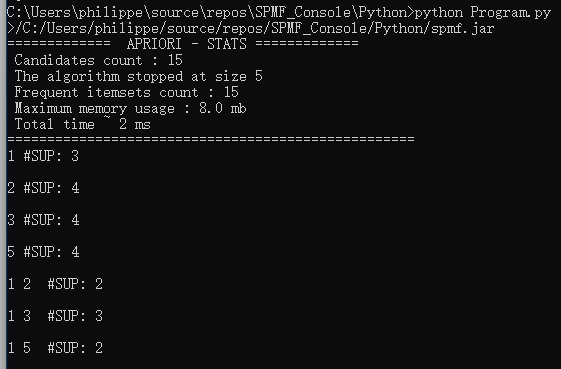
Everytime that the user calls an algorithm from either the command line or graphical interface, the method runAlgorithm() of the Command Processor is called as illustrated below:
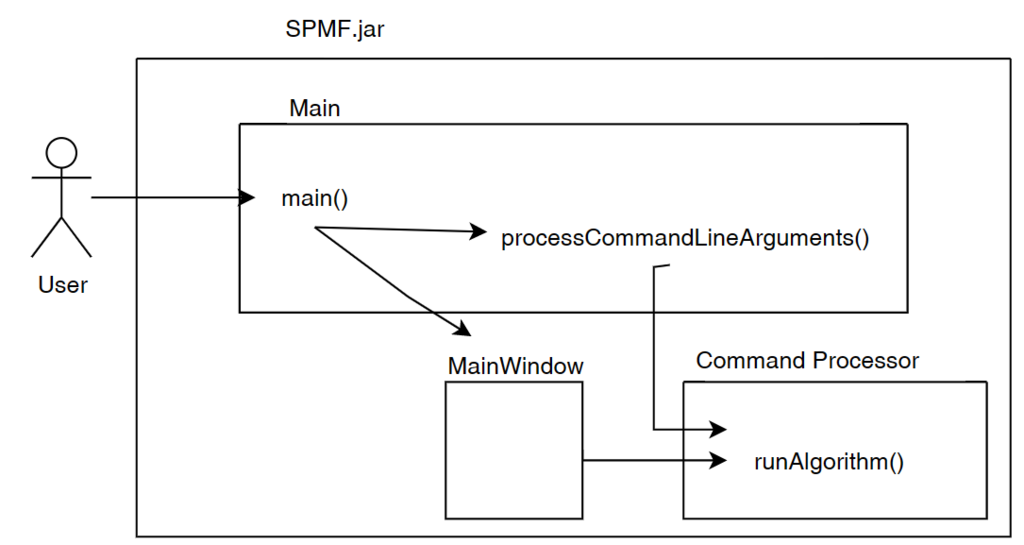
The runAlgorithm() method of the command processor takes as parameters: (1) an algorithm name, (2) a path to an input file (or null), (3) a path to an output file (or null), and (4) an array of parameters to be passed to the algorithm. Here is the declaration of this method:

What does the Command Processor do when the runAlgorithm() method is called? It does the following:
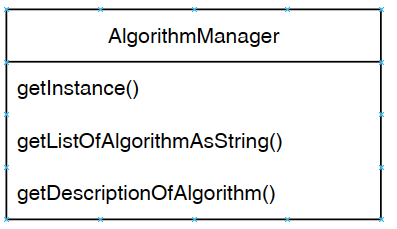
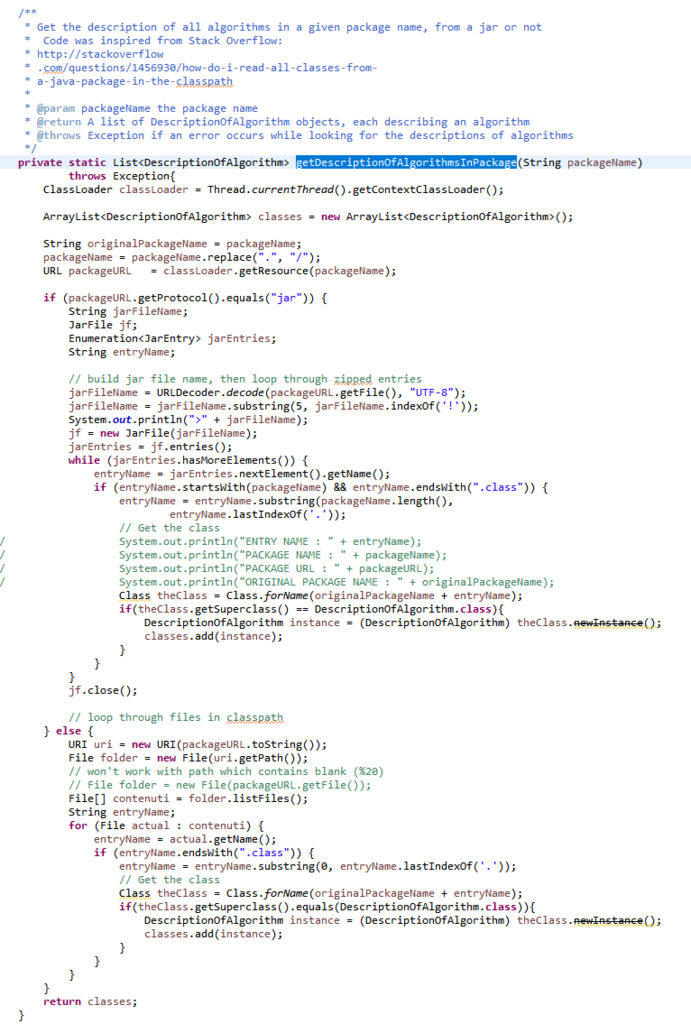
First, the Command Processor calls the method getDescriptionOfAlgorithm() of the Algorithm Manager to obtain information about the algorithm that the user wants to run, as illustrated below.
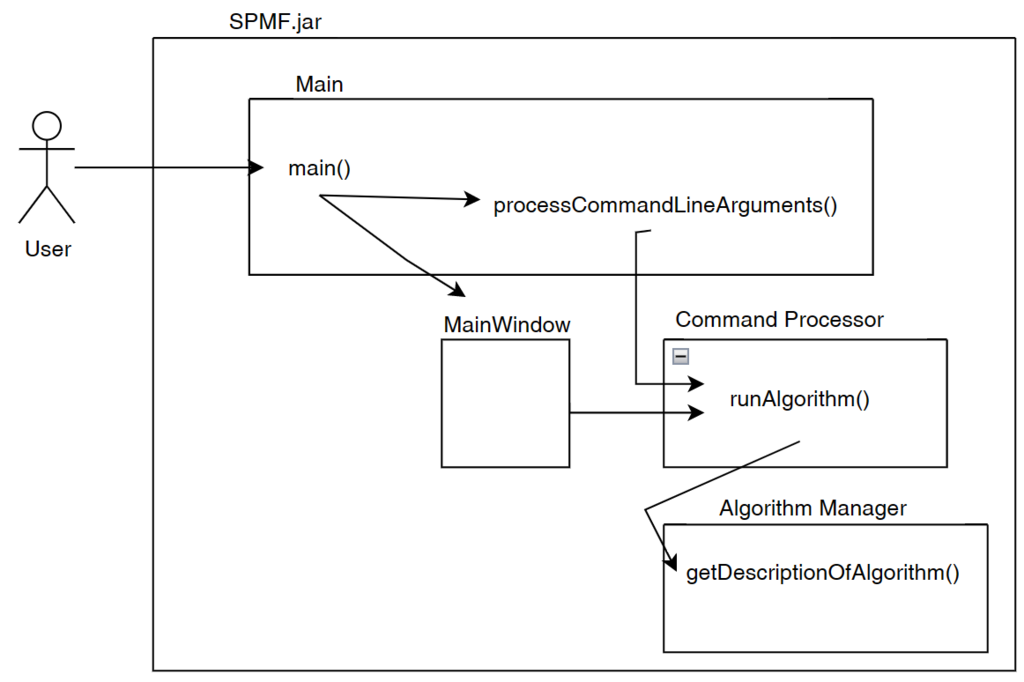
After obtaining information about the algorithm, the Command Processor compares this information to the parameters provided by the user. If the algorithm does not exist in SPMF, if the input or output file path are incorrect, or if the parameters provided by the user do not match the description of the algorithm, an error is thrown. This error will be displayed to the user either through the graphical or command line interface.
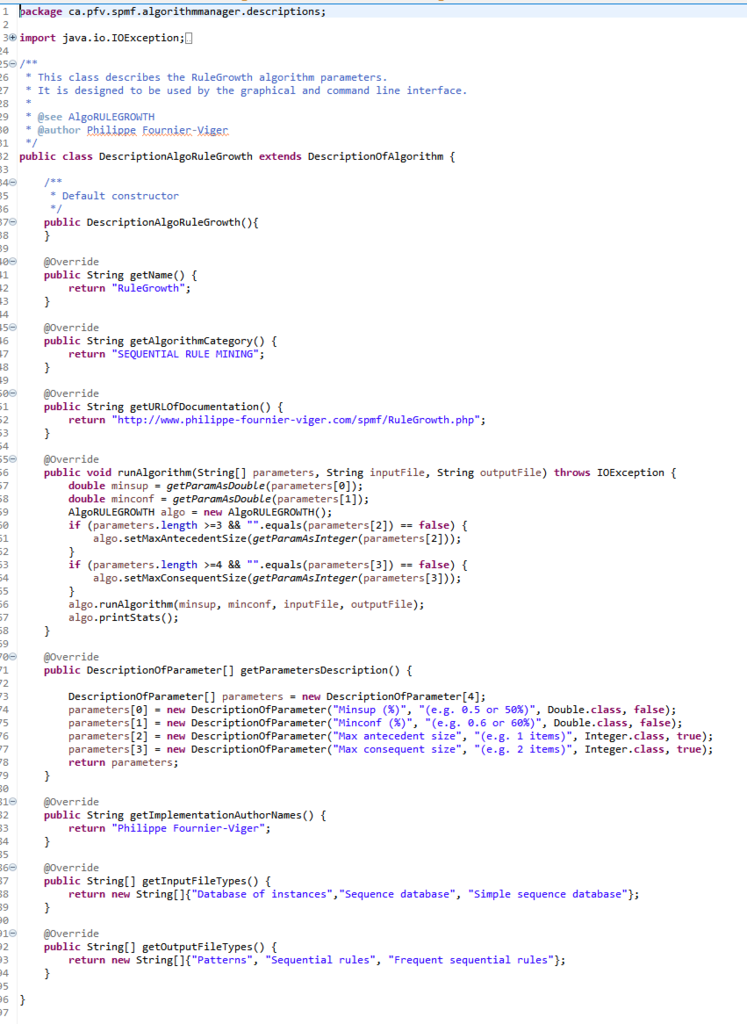
After that, the Command Processor will run the algorithm() by calling the algorithm based on its description. The description of an algorithm is a subclass of the class DescriptionOfAlgorithm and must have a runAlgorithm() method. The Command Processor call this method to run the algorithm. This is illustrated below:
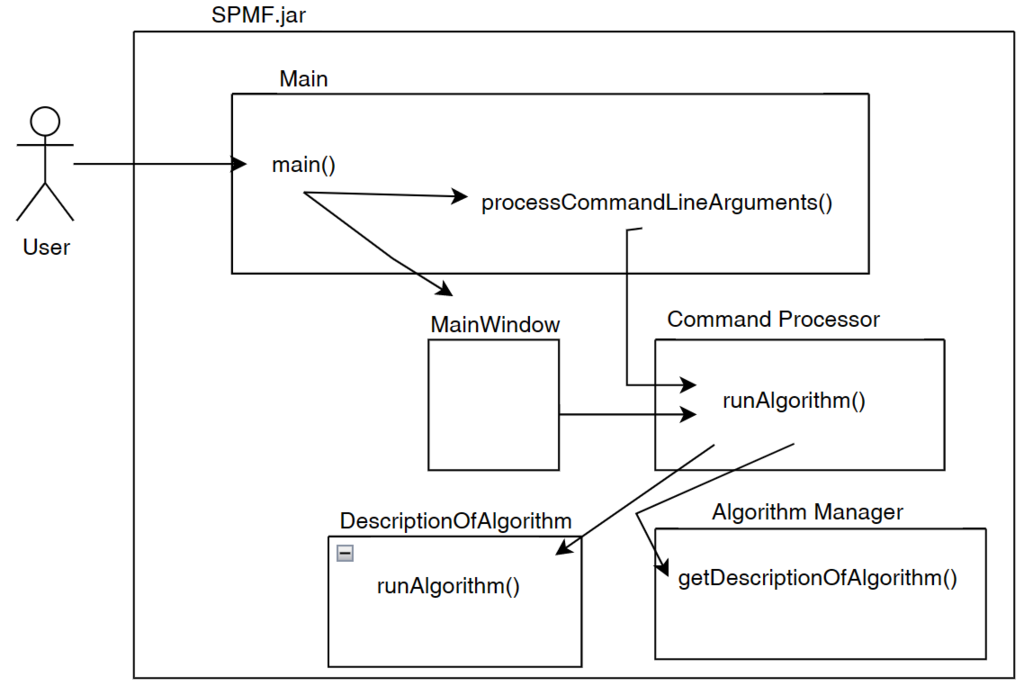
So until now, I have explained the main idea about the Main class and the Command Processor. Now, I will explain a bit more details.
The Command Processor can also automatically convert some file format
Another feature of the Command Processor is that it can automatically convert some special file formats to the SPMF format so that SPMF algorithms can be run on other file formats and that it is totally transparent to the user, and that algorithms dont need to be modified to support other formats.
This is achieved as follow. If the Command Processor is called with a special file type such as files having the extensions .ARFF or .TEXT files, then the Command Processor will call some tools to convert these files to the SPMF format. This will produce some temporary file. Then the Command Processor will call the requested algorithm on this temporary file. And finally, the Command Processor will delete the temporary file, and convert the output of the algorithm back to the format requested by the user. I might explain this in more details in a future blog post.
A more accurate picture of SPMF’s architecture
So after what I have explained today, we can get a more clear picture of the architecture of SPMF as follows, where I have added the Main class and the Command Processor:
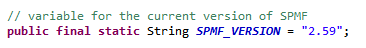
SPMF version number
By the way, the Main class is also where the version number of SPMF is stored in the code:
Conclusion
In this blog post, I have explained more about the architecture of SPMF. In particular, I have described the role of the Main class and the Command Processor, which are key to run SPMF both from a command line interface and graphical interface.
Hope that it has been interesting. If so, please leave some comments below 🙂
==
Philippe Fournier-Viger is a distinguished professor and the founder of the open-source data mining software SPMF, offering more than 250 data mining algorithms.