
Just a short blog post today to wish you happy holidays and Merry X-Mas to those who are celebrating it, among the users of SPMF! Thanks again for your support!

Just a short blog post today to wish you happy holidays and Merry X-Mas to those who are celebrating it, among the users of SPMF! Thanks again for your support!
Today, the first workshop 1st Workshop on Heuristic and Pattern Mining for Multi-Omics Data Analytics was held at IEEE BIBM 2025, online. I co-organize this workshop with M. Saqib Nawaz and other collaborators. The workshop focus on various machine learning and pattern mining methods and their applications to the analysis of multi-omics data.
Here are the slides from the opening ceremony:

It was announced that 8 papers have been accepted by the workshop this year. They cover multiple topics such as interpretable deep learning for regulatory sequence analysis, graph neural networks for single-cell and spatial omics data, heuristic and bio-inspired optimization for protein fitness landscapes, minimum description length and evolutionary approaches for protein compression, advanced sequential and temporal pattern mining methods, and multi-source data fusion models for predicting complex genetic traits.
We also mentioned that we are currently working to organize a special issue for extensions of the papers (to be confirmed later).
We also announced that the best paper award of the workshop was given to this paper by researchers from Canada:
Mahshad Hashemi, Sharjeel Mustafa, Alioune Ngom, and Luis Rueda, HeteroGraphNet: A Ligand–Receptor Informed, Heterophily-Adapted Graph Neural Network for Cell Type Prediction in scRNA-seq Data
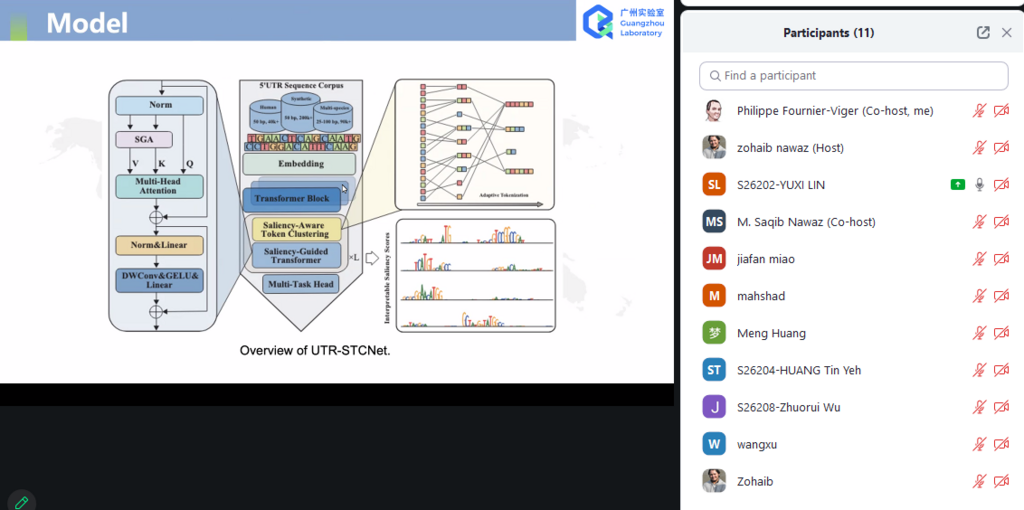
There was several interesting presentations on diverse topics. Here for example, a screenshot from the first paper presentation by Lin, Yuexi et al. about Decoding Translation-Related Functional Sequences in 5’ UTRs Using Interpretable Deep Learning Models:
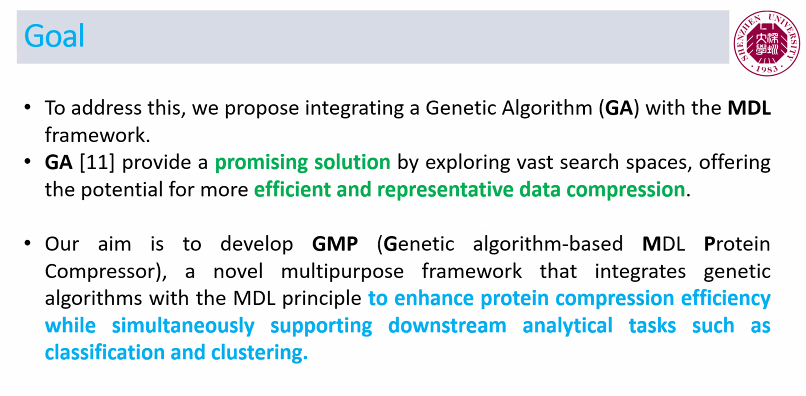
Among the papers, my PhD student presented a new algorithm called GMP for protein sequence compression based on pattern mining. Here are a few slides to show an overview:
This is just a short report about the workshop. It has been a success for the first edition of this workshop. We thus plan to organize it again next year!
In November, I released the version 2.64 of SPMF with 5 new algorithms and 2 new tools, among other improvements, but I still had several other pattern mining algorithms waiting to be released after more tests.
Thus, today, I make a new release of SPMF for December 2025, called version 2.64b. This release has several new algorithms:
Moreover, there is a new Pattern Diff Analyzer tool that allows to compare two pattern files to find contrast patterns. This tool works letting the user first select two files containing patterns:
And then we can compute the differences between these files using different contrast methods. For example, we can identify all patterns that are in the first file but not in the second, among multiple other options for identifying contrast patterns:
That is all I wanted to share about this new version! Hope that you will enjoy this new release of the SPMF pattern mining software. So far we have released 18 new algorithms this year! And more features are coming soon…
Recently, I have been working on improving the SPMF data mining software. Something good about SPMF is that it has a simple user interface. But as SPMF has evolved with more and more algorithms, the list of algorithms in the software has become very long and it may be not so easy to browse through the list of algorithms. Thus, I have started to think about upgrading the user interface to make it more user-friendly. Here is some new prototype welcome window for SPMF that I am working on:
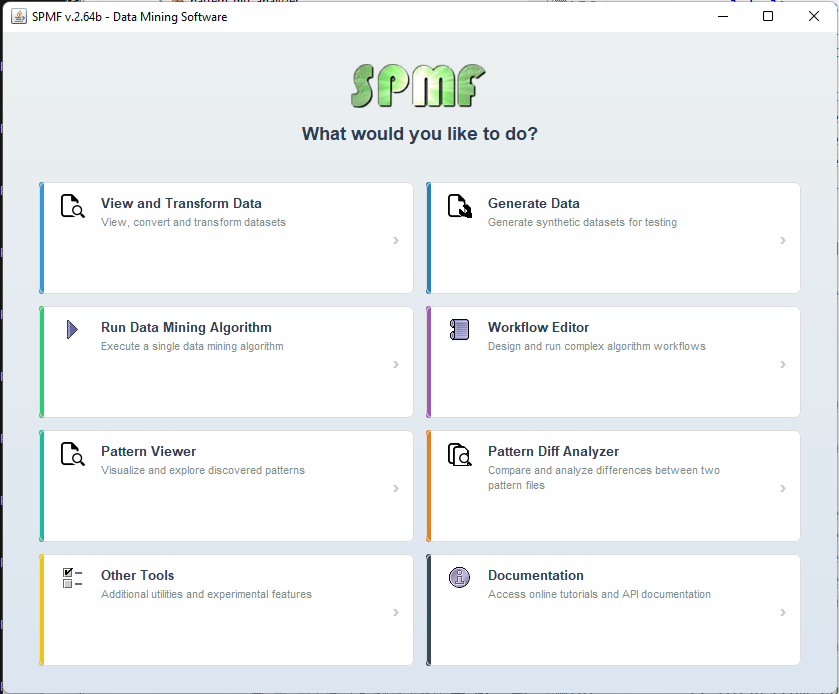
This window provides access to all the main features of SPMF through a centralized screen. Thus, the user can clearly focus on the different tasks such as generating data, or choosing a data mining or pattern mining algorithm, or viewing and transforming data. When the user will click on “View and transform data” for example, he will access only the algorithms and tools for viewing and transforming data.
I think that this type of interface can be an improvement over the existing user interface. However, for now, this is only a prototype and I am working on putting this all up together, and testing. I will not release a new interface for SPMF until I am sure that everything works well and that it is good. And I might also leave the option of choosing between the traditional user interface and the new user interface.
If you have any ideas or suggestions to make this better, please leave me a comment below or email me! I think that if work on this user interface go well, maybe it could be released early next year.
Again, thanks for all users of the SPMF pattern mining library for your support!
Today, I will talk to you about an upcoming feature of SPMF pattern mining software 2.64b, which I think will be very useful to many people. It is a new tool, called the Pattern Diff Analyzer that allows to calculate the contrast between two files containing patterns.
For example, lets say that you extract sequential patterns from two text documents. You can now use this tool to find variations in the patterns found in both document to discover patterns that distinguish each document. Another example is you extract patterns from the genome sequences of two viruses and want to find patterns that differ in the two sequences.
The new Pattern Diff Analyzer tool is very simple to use and looks like this:
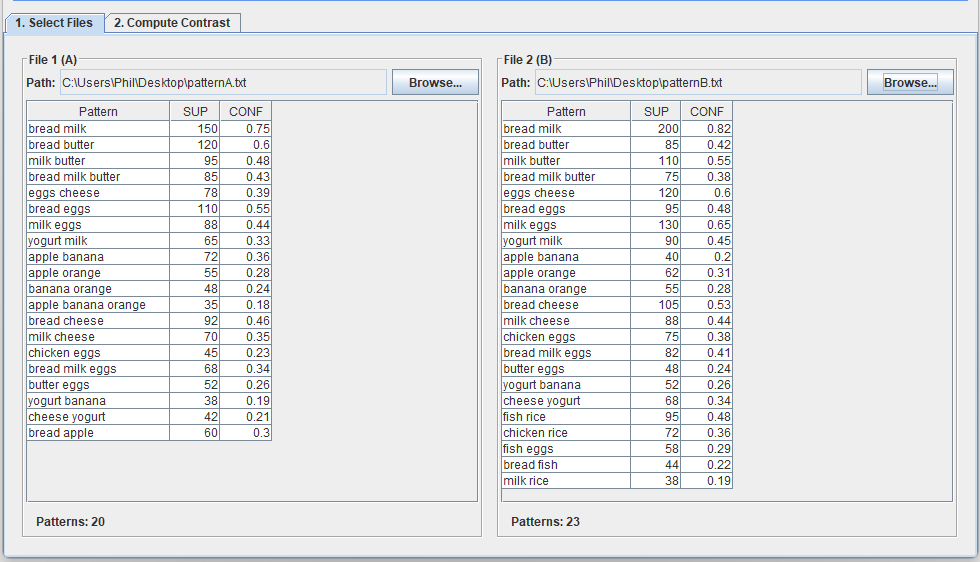
In this screen, we can select two files containing patterns found by a pattern mining algorithm. For example, I will use two files called patternsA.txt and patternsB.txt.

After that, we can go to the second tab called “Compute contrast” to find the differences in patterns between these two files. In the picture below, I choose the “SUP” measure (support) for calculating the difference, and I choose “Absolute difference” with the threshold of 10. This means that I want to find all the patterns where the difference in support is more than 10 between the two files. The result is 20 patterns:
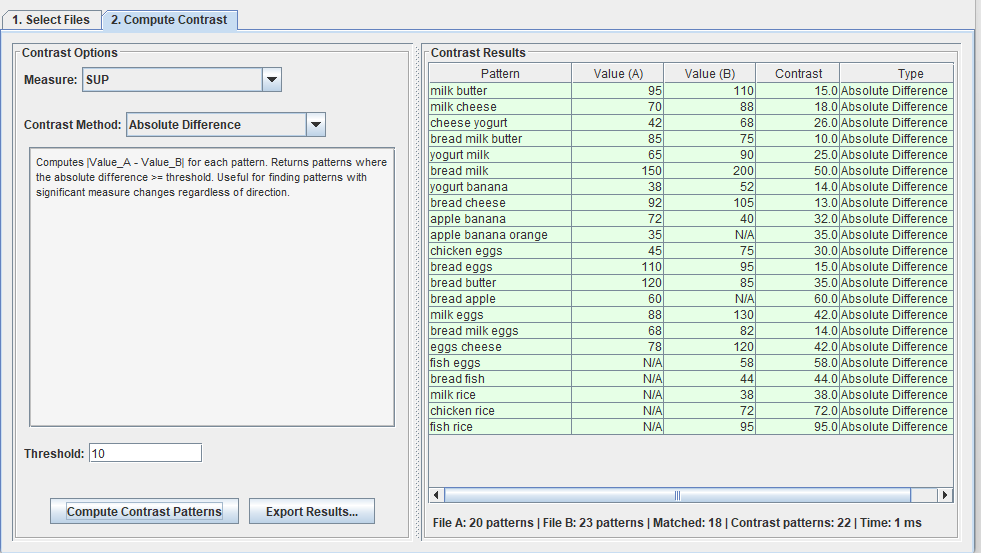
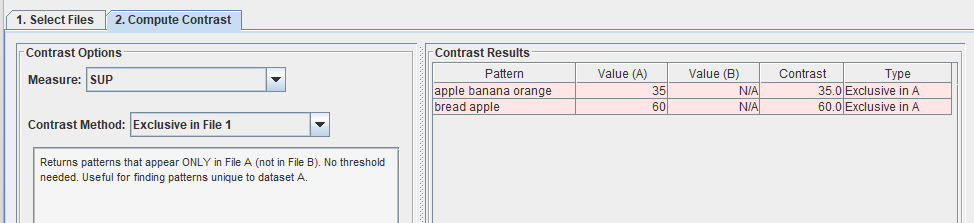
I can also choose other contrast methods such “Exclusive in file 1“, which means all patterns that only appear in the first file but not in the second file:

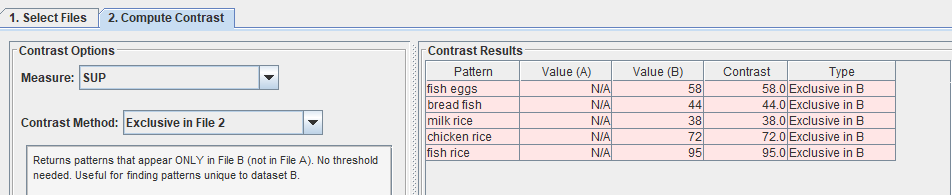
Or similarly, I can choose “Exclusive in file 2“:
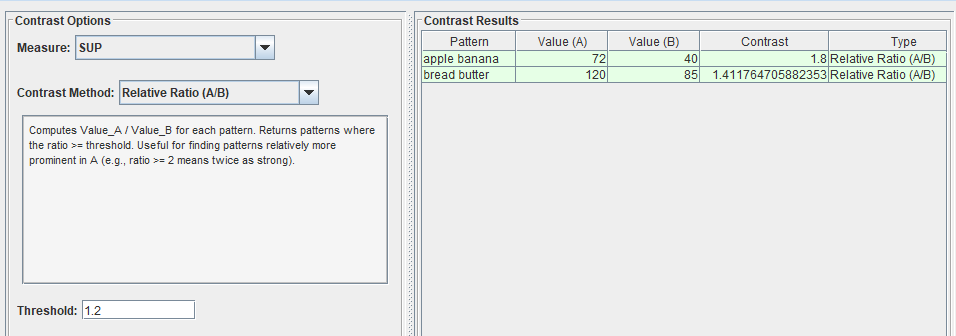
There are also other contrast methods available such as the ratio of a pattern’s measure value for file A to that in file B. For example, here I select patterns where the ratio of A to B is at least 1.2:
After discovering the contrast patterns, we can also Export them to a text file for saving these results!
I think this tool will be very useful for classification problems where we want to compare patterns from different classes. Related to classification, note that in SPMF, we also have multiple algorithms for classification using association rules. But this is a different approach.
So today, I just wanted to show you a preview of this upcoming tool in SPMF. I will continue testing and may made some changes before the final release. Also, I will provide an algorithm that could be called from the command line to do the same thing as this Pattern Diff Analyzer tool so that it can be used without the graphical user interface as well.
Today, I want to talk to you about something quite incredible. I recently attended a keynote speech at an international conference. The keynote speaker was presenting online and was scheduled for a 35 minute presentation. However, he had prepared 180 slides. Yes, you have read that correctly: 180 slides!
This is a ridiculous number of slides for a talk. Generally, a good rule of thumb for a presentation is to prepare one slide per minute. If we follow this rule, a 35 minute presentation should have 35 slides. So, as I would expect in such case, the talk went overtime…
After 1 hour (already 25, the speaker was at slide 140, while going at a speed of 2.3 slides per minute!
Then, a few minutes later, the organizer intervened to stop the talk. And the speaker said sorry for going overtime…. But seriously, how can you be sorry when you prepare 180 slides.
I have seen some speaker exceed the time allowed at several conferences. However, usually, it is by 5 or 10 minutes, rarely by over half an hour. And if he had not been stopped, at this speed, it would have taken almost 1 hour and half to finish the presentation.
Personally, I do not understand this. If you are invited to give a talk at a conference, I think it is important to follow the schedule, so as to show respect to the organizers, to the audience and to the speakers that will talk after you. Because of that speaker, other speakers had to shorten their presentations, and the coffee break had to be shortened, as well.
What’s more, it seems that the professor was well aware that he had too many slides as the title of his presentation was ironically titled “The too short keynote speech of Prof (…. ) “, as you can see in the picture below:
I will not say the name of the speaker. But he is a senior Jordanian professor with over 20,000 citations.
So my goal by telling this story is just to remind everyone that it is important to follow the time constraints at conferences. It shows respect and professionalism. That is all I wanted to say for today! If you want to share your opinion, please leave a comment below.
Today, I am happy to announce that a new version of the SPMF data mining library has been released.
It introduces five new algorithms:
Besides, some various other improvements have been made to the SPMF software, including a new tool in the GUI of SPMF called the Algorithm_Graph_Visualizer to visualize the similar algorithms in terms of input and output or categories as a graph (I had discussed about that in a recent blog post):
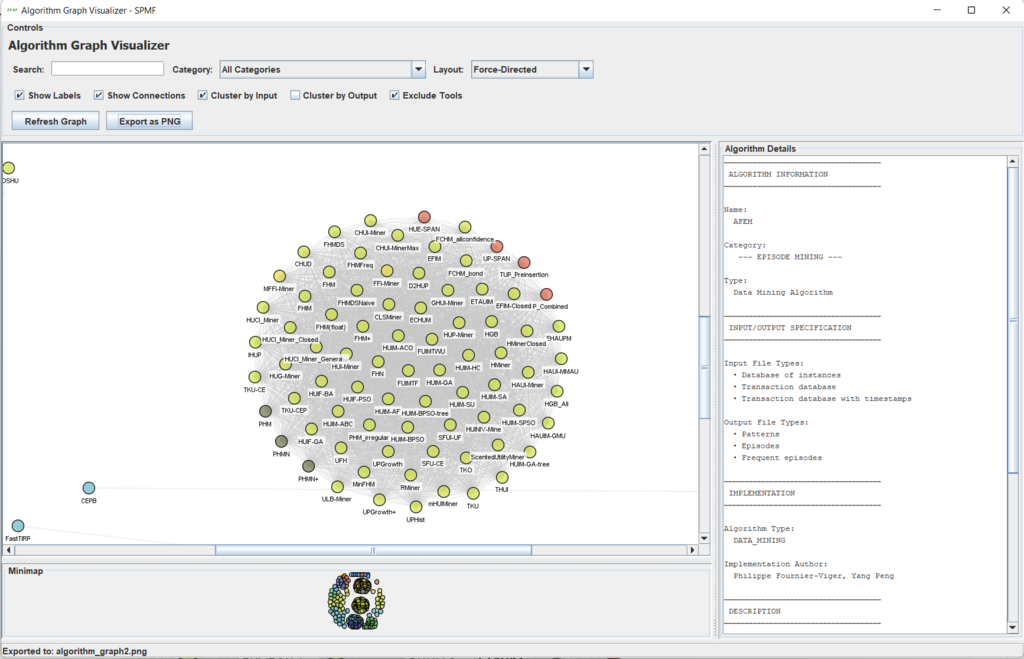
Besides, I have improved various graphical interface components such as the MemoryViewer, HistogramViewer, GraphViewer and SPMF Text Editor by fixing a few bugs and adding new features. I have also added a new mode in the TransactionDatabaseViewer to view transactions either as lists or as columns.
If everything goes, I might release more algorithms before the end of this year or early next year. I have several algorithms waiting to be integrated in SPMF!
To all users, thanks again for using the software! And to all contributors: thanks for your help!
If you have any algorithm that you would like to integrate in SPMF, feel free to contact with me by e-mail.
Today, I am pleased to share that our team has published a new algorithm called GMP for the compression and analysis of protein sequences. The paper will appear next month at BIBM 2025. Here is the full reference:
Nawaz, M. Z., Nawaz, S., Fournier-Viger, P, Niu, X., Li, M. (2025). A Multipurpose Protein Compressor based on MDL and Genetic Algorithm. Proceedings of BIBM 2025.
And you can watch the video of the presentation on Youtube:
Presentation (18 minutes)
Abstract
The rapid expansion of protein sequence databases has created challenges for efficient storage, transmission, and analysis. Unlike genomic sequences with only four nucleotide bases, proteins are composed of twenty amino acids, making compression more complex. Existing specialized protein compressors, such as AC, AC2, and CPM-FCM, have achieved promising performance but still face limitations, including high computational cost, low adaptability, and limited biological interpretability. This paper introduces GMP (Genetic algorithm-based MDL Protein compressor), a novel protein compression framework that leverages the Minimum Description Length (MDL) principle with a genetic algorithm to discover optimal patterns of amino acid subsequences (kAA-mers). Experimental results demonstrate that GMP attains compression performance comparable to state-of-the-art methods while additionally supporting tasks such as classification and clustering—capabilities absent from traditional protein compressors. This makes GMP not only an efficient compression framework but also a biologically interpretable tool for protein sequence analysis. GMP is available at github.com/MuhammadzohaibNawaz/GMP.
Index Terms—Protein sequences, Compression, Genetic Algorithm, Minimum Description Length, kAA-mers
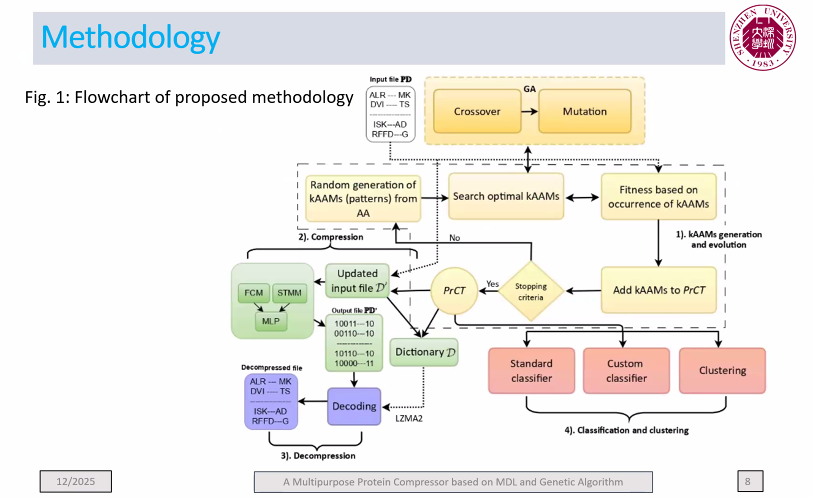
GMP was designed not only to compress protein sequences but also to provide insights into their structure through the discovery of meaningful subsequence patterns. By integrating MDL with a genetic algorithm, it strikes an effective balance between compression quality and interpretability. One of the unique strengths of GMP is that it can simultaneously serve multiple purposes: compression, classification, clustering, and pattern discovery—functions rarely combined in a single framework. Here is the main flowchart from the paper:
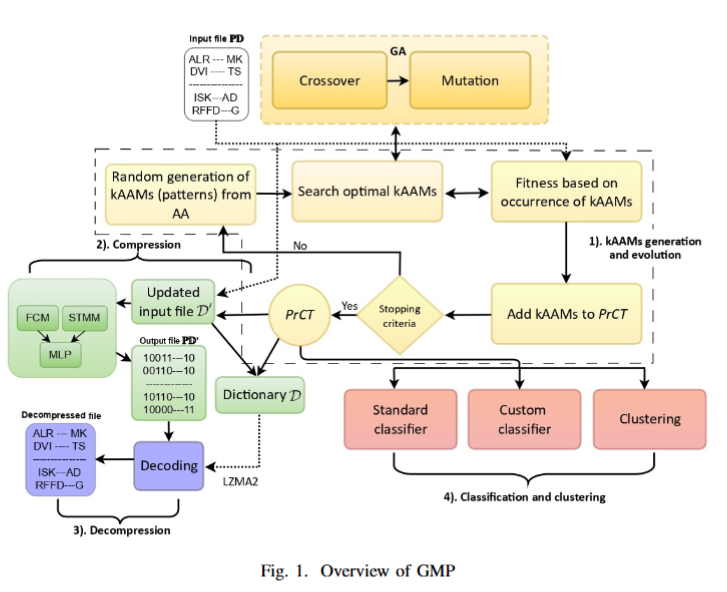
We will release the paper soon after it is published next month at BIBM 2025.
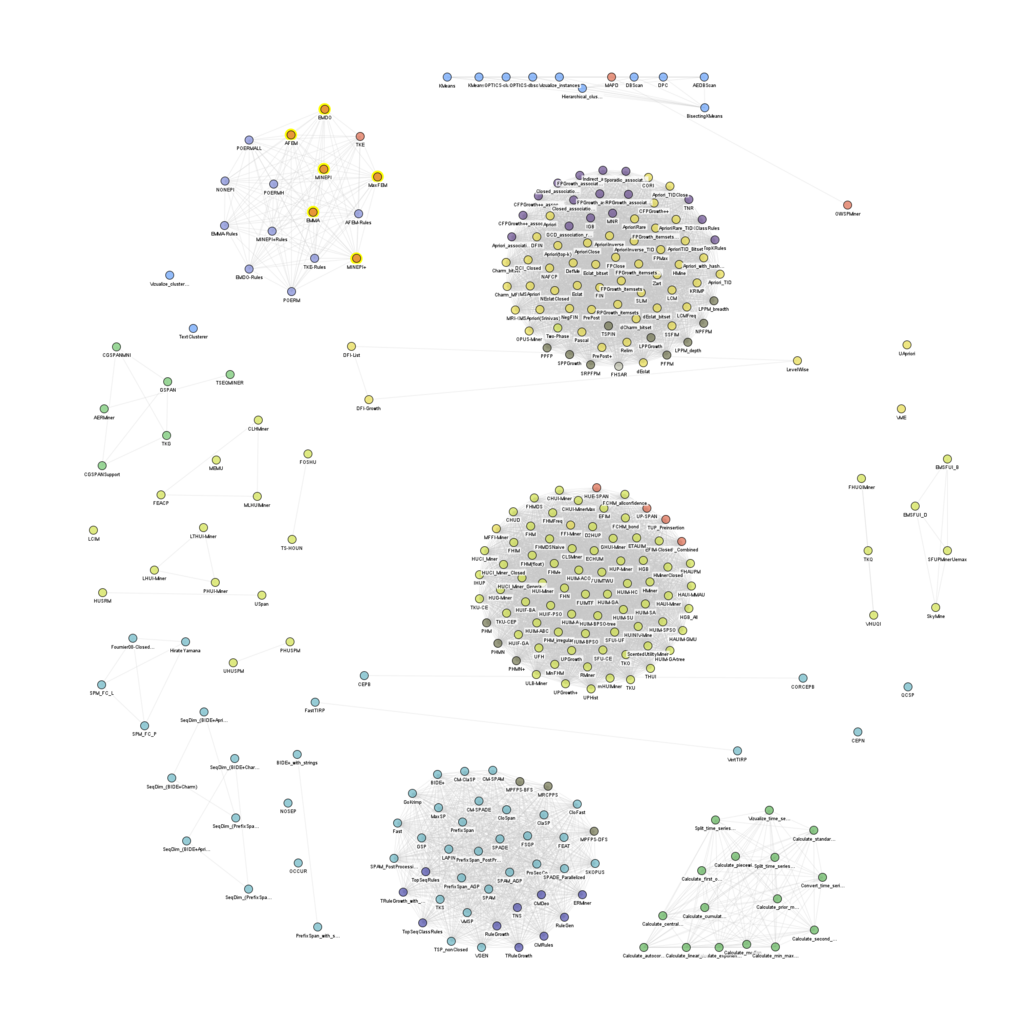
Today, I wrote some code to visualize the relationships between the algorithms offered in the SPMF open-source pattern mining library. Here is the graph of all algorithms in SPMF (excluding tools) that have the same input and output type:
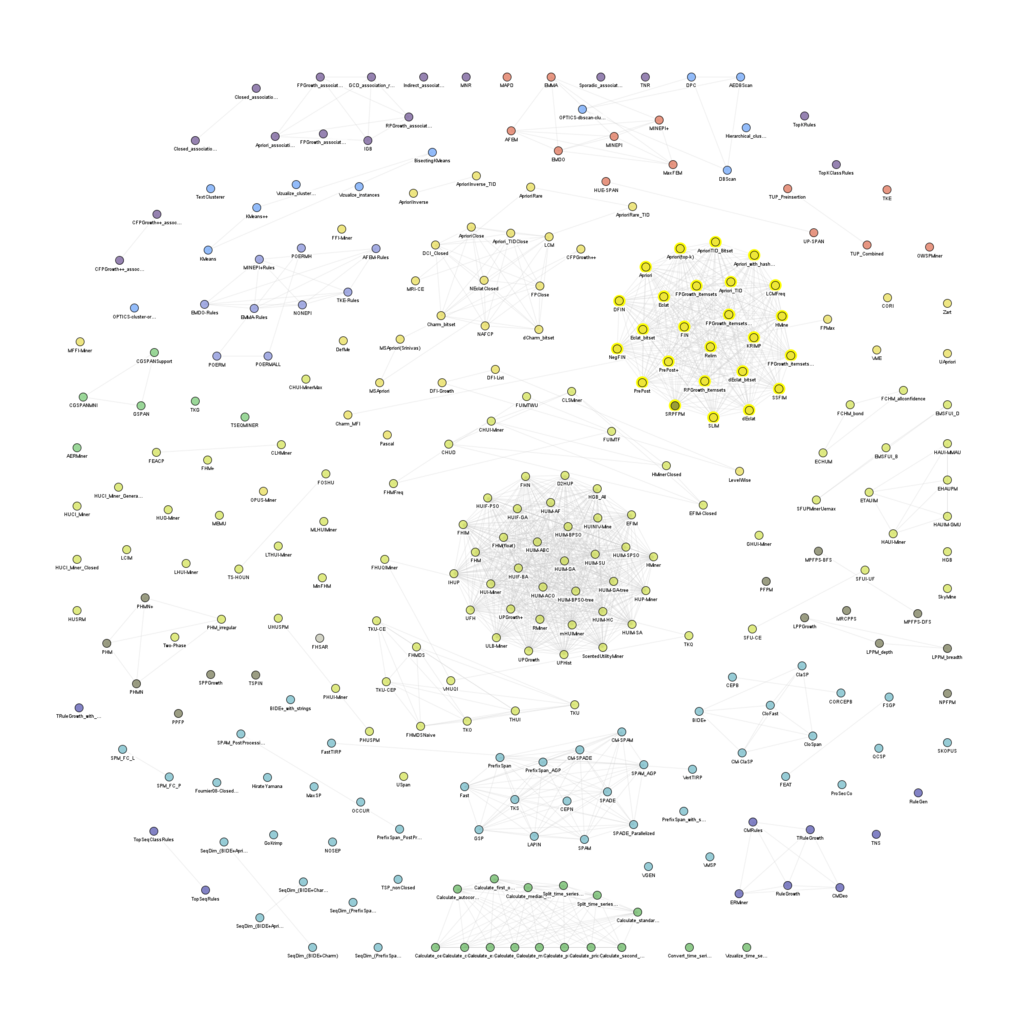
We can see a few big clusters such as high utility itemset mining algorithms:
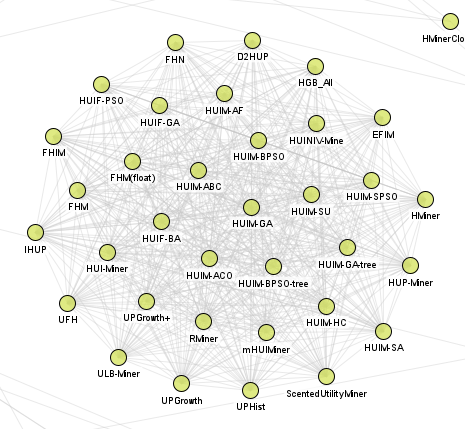
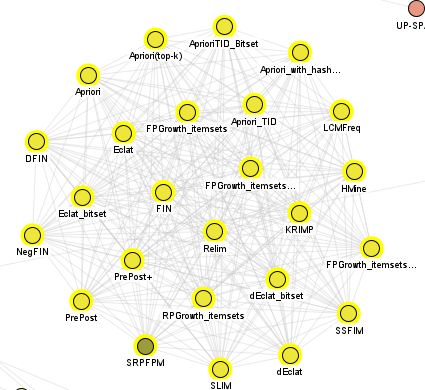
Frequent itemset mining algorithms:
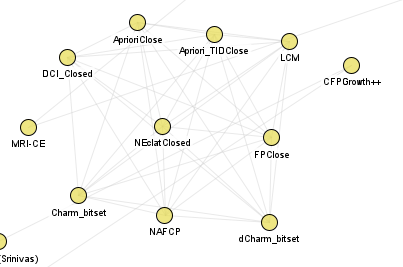
Closed itemset mining algorithms:
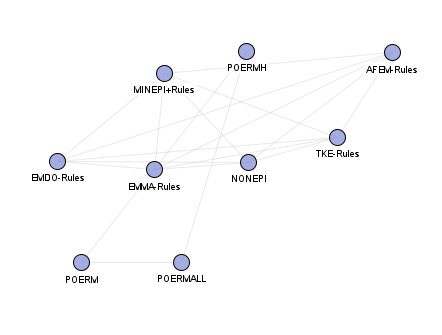
Episode rule mining algorithms:
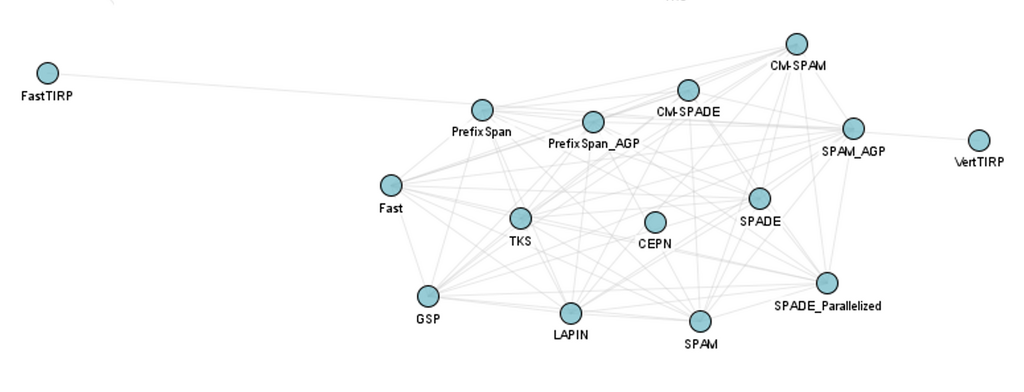
Frequent sequential pattern mining algorithms:
Sequential rule mining algorithms:
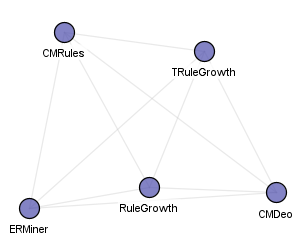
Frequent episode mining algorithms:
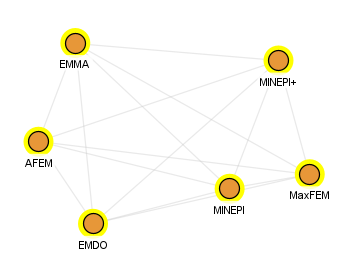
If we only display the algorithms using edges if they have the same input file type (but may not have the output), the graph is different:
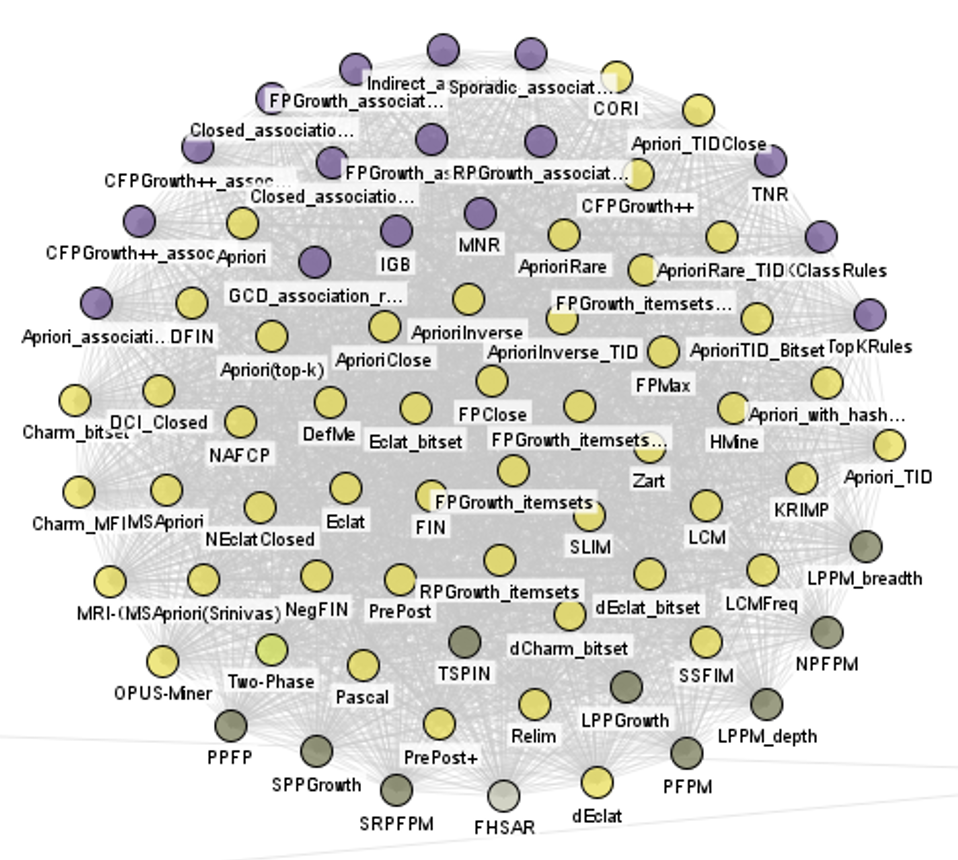
Now, there is a huge cluster for itemset mining and association rule mining:
And there is a big cluster for sequence mining algorithms:
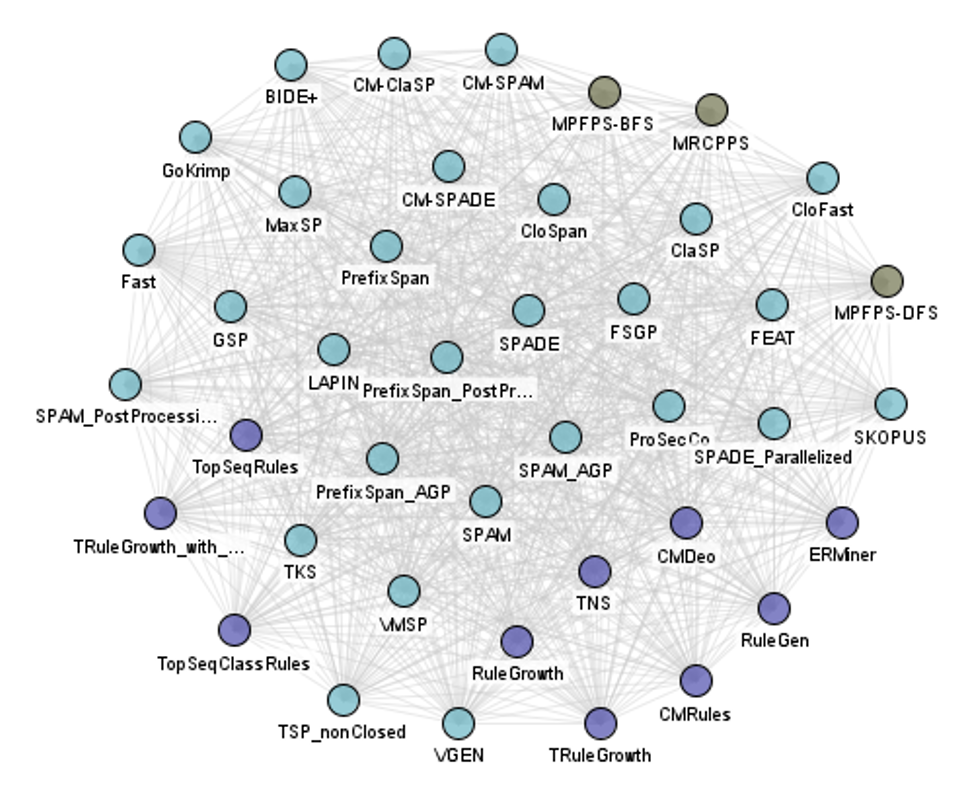
And a smaller cluster for episode mining:
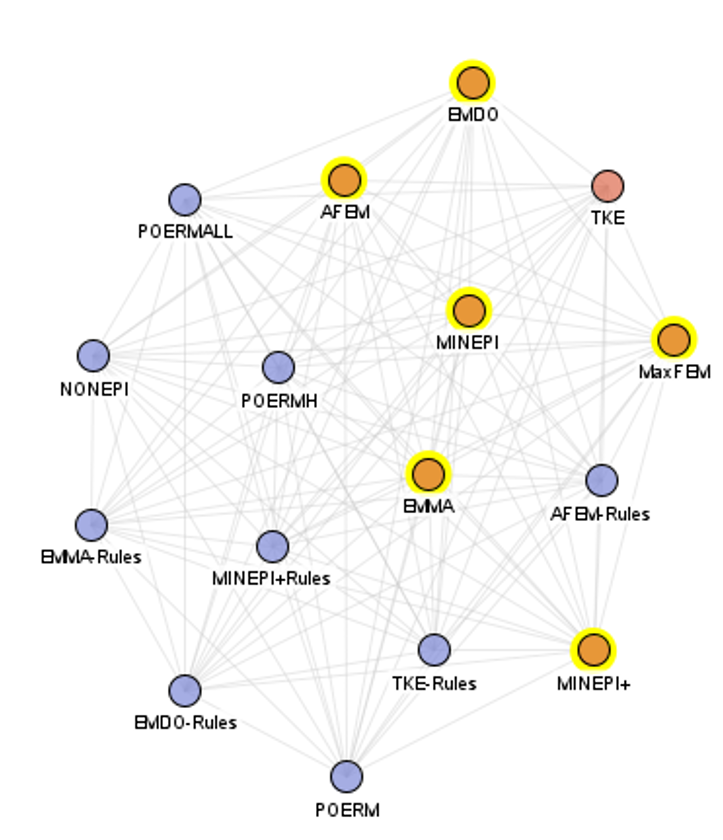
and a big cluster for high utility pattern mining:
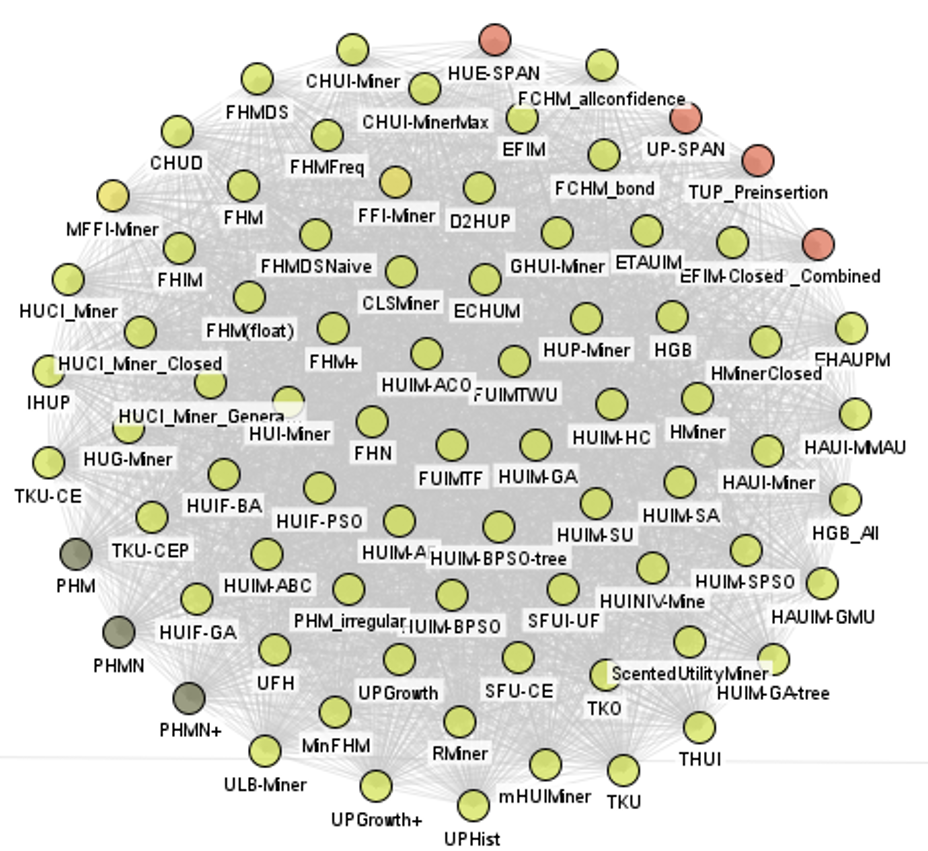
The tool that I use to draw these pictures is the AlgorithmGraphVisualizer, a new GUI tool, which will be offered in the next version of SPMF. It allows to visualize algorithm relationships with different options and export to PNG. Here is the current GUI interface:

Hope that this has been interesting! The new version of SPMF should come out probably next week!
Today, I will show how to fix the Latex reviewresponse.cls class to allow multi-page comments.
If you have ever written a detailed response to reviewers in LaTeX, you may have noticed that long reviewer comments sometimes get cut off instead of continuing on the next page. This happens because the comments are enclosed in non-breakable tcolorbox environments.
In the original version of reviewresponse.cls, the environments for reviewer comments look something like this:
\newenvironment{generalcomment}{%
\begin{tcolorbox}[attach title to upper,
title={General Comments},
after title={.\enskip},
fonttitle={\bfseries},
coltitle={colorcommentfg},
colback={colorcommentbg},
colframe={colorcommentframe},
]
}{\end{tcolorbox}}
\newenvironment{revcomment}[1][]{\refstepcounter{revcomment}
\begin{tcolorbox}[adjusted title={Comment \arabic{revcomment}},
fonttitle={\bfseries},
colback={colorcommentbg},
colframe={colorcommentframe},
coltitle={colorcommentbg},
#1
]
}{\end{tcolorbox}}
\newenvironment{changes}{\begin{tcolorbox}[colback={colorchangebg},
colframe={colorchangeframe},enhanced jigsaw,]
}{\end{tcolorbox}}These definitions produce nice colored boxes, but the problem is that tcolorbox by default does not break across pages. When your reviewer writes a long paragraph, LaTeX tries to keep the entire box on one page, which can result in missing text or strange layout issues.
The Solution
The fix is simple: you need to make the boxes breakable and enhanced. The tcolorbox package provides two key options for this:
breakable — allows the content to flow onto the next page.enhanced jigsaw — ensures compatibility with decorations, titles, and other layout features when breaking boxes.Here is the fixed version of the environments:
\newenvironment{generalcomment}{%
\begin{tcolorbox}[
enhanced jigsaw,
breakable,
attach title to upper,
title={General Comments},
after title={.\enskip},
fonttitle={\bfseries},
coltitle={colorcommentfg},
colback={colorcommentbg},
colframe={colorcommentframe},
]
}{\end{tcolorbox}}
\newenvironment{revcomment}[1][]{%
\refstepcounter{revcomment}
\begin{tcolorbox}[
enhanced jigsaw,
breakable,
adjusted title={Comment \arabic{revcomment}},
fonttitle={\bfseries},
colback={colorcommentbg},
colframe={colorcommentframe},
coltitle={colorcommentbg},
#1
]
}{\end{tcolorbox}}
\newenvironment{revresponse}[1][{}]{%
\textbf{Response:} #1\par
}{\vspace{4em plus 0.2em minus 1.5em}}
\newenvironment{changes}{%
\begin{tcolorbox}[
enhanced jigsaw,
breakable,
colback={colorchangebg},
colframe={colorchangeframe},
]
}{\end{tcolorbox}}
Result
After this modification, your reviewer comments and “changes” boxes will automatically continue onto the next page, no matter how long they are. You can now safely include large comments or detailed explanations without worrying about text being cut off.
Conclusion
By simply adding enhanced jigsaw and breakable to the tcolorbox environments, you make your LaTeX review responses much more robust. This small fix prevents truncated comments and keeps your document professional and reviewer-friendly.