Recently, my team has been working on analyzing COVID-19 genome sequences using pattern mining and other data mining and AI techniques. We have recently published a paper in the Applied Intelligence journal about this.
In this blog post, I will give some brief overview of this. The PDF of the paper can be found here:
Nawaz, S., Fournier-Viger, P., Shojaee, A., Fujita, H. (2021). Using Artificial Intelligence Techniques for COVID-19 Genome Analysis. Applied Intelligence, to appear. |
And the source code and data: https://github.com/saqibdola/SPM-MA4GSA
The main idea of the paper is the following. We have obtained genome sequences of different strains of the COVID-19 virus. These genome sequences can be viewed as strings of letters (nucleotides). For example, below is four sequences of nucleotides:
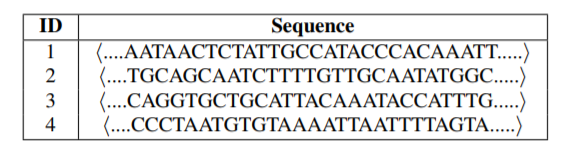
Then, after preprocessing these sequences, it is possible to analyze them using pattern mining algorithms and other artificial intelligence techniques. The main process is the following:
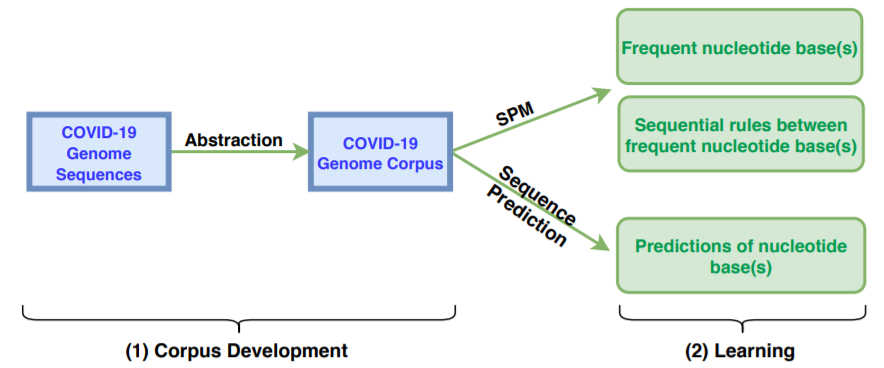
First we prepare the data (step 1), and then we apply different techniques (step 2). First, we have applied itemset mining, sequential pattern mining and sequential rule mining techniques to find patterns that are common to many genome sequences. Some examples of sequential patterns (sequences of nucleotides that appear often) find by the CM-SPAM algorithm are below:
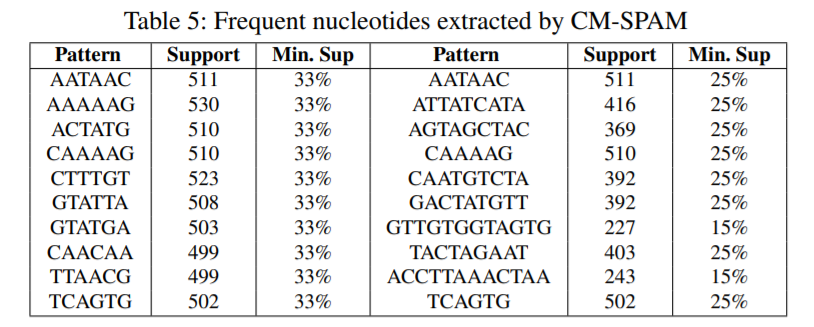
This is just to give an overview. Other types of patterns are discussed in the paper in more details.
Second, we also tested sequence prediction models to see if the nucleotides in genome sequences can be predicted. We compared various models offered in the SPMF data mining software and got results like this:
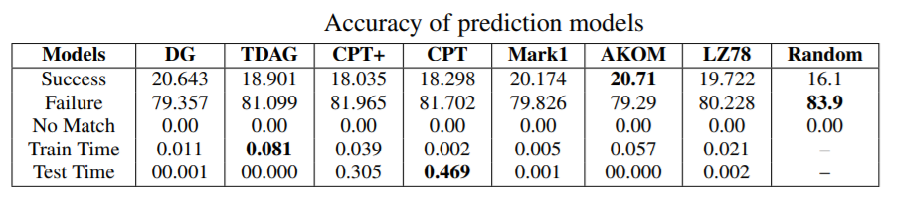
In general, prediction of genome sequence does not give a high accuracy but still better than a random prediction. We discuss these results in more details in the paper.
Third, we also designed some mutation analysis algorithm to compare different strains of the coronavirus. For example, by comparing two strains, we identified some mutations:
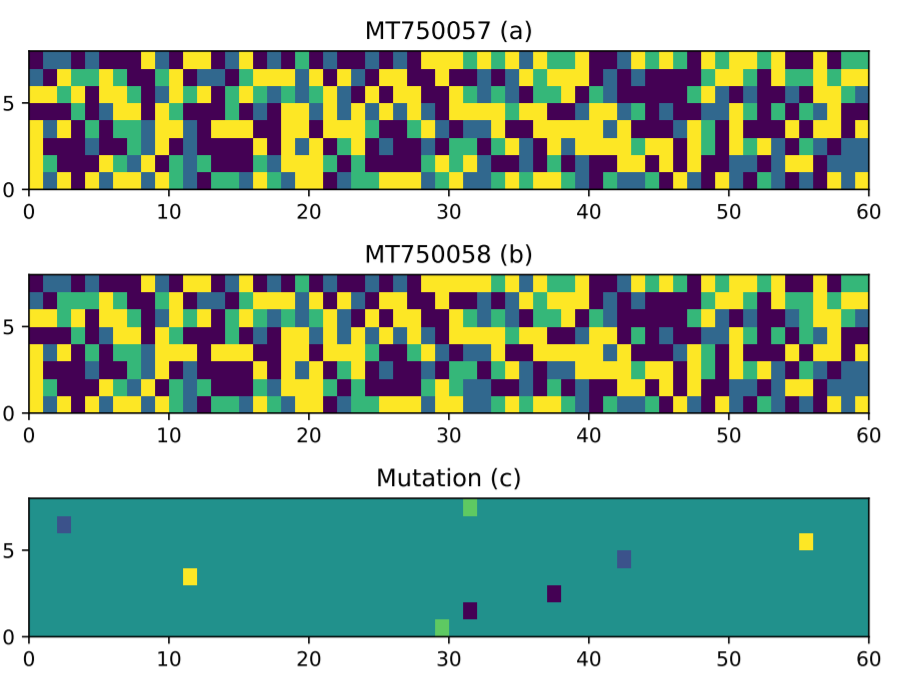
That is just a brief overview of the paper!
There are many possibilities for extensions of this research. In particular, various other pattern mining algorithms and machine learning algorithms could be applied as well. Using the code and data provided above, you can also make your own research on this topic! Besides, the tools presented in this paper can also be applied to other genome sequences beside the COVID-19 virus.
Hope this has been interesting.
—
Philippe Fournier-Viger is a professor of Computer Science and also the founder of the open-source data mining software SPMF, offering more than 120 data mining algorithms.






Good idea Prof. Fournier-Viger
Bay
Hi Bay!
Thanks for your comment on the blog. Happy to see you there 🙂
Best,
Philippe
Pingback: An Overview of Pattern Mining Techniques (by data types) | The Data Mining Blog
Prod. Philippe your research ideas, papers gives basic knowledge for my research and future findings.
Thank you so much Prof.
Thanks for reading the blog. Happy it is inspiring you 🙂