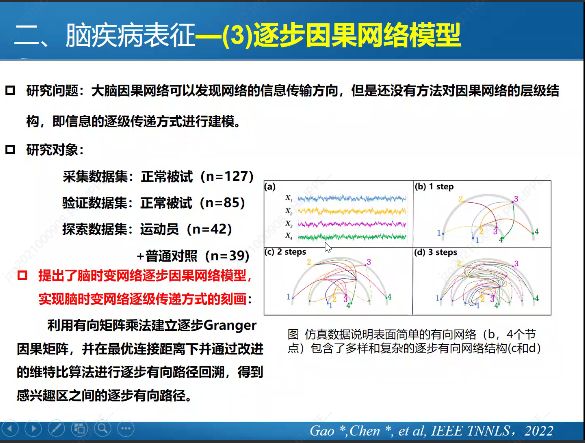
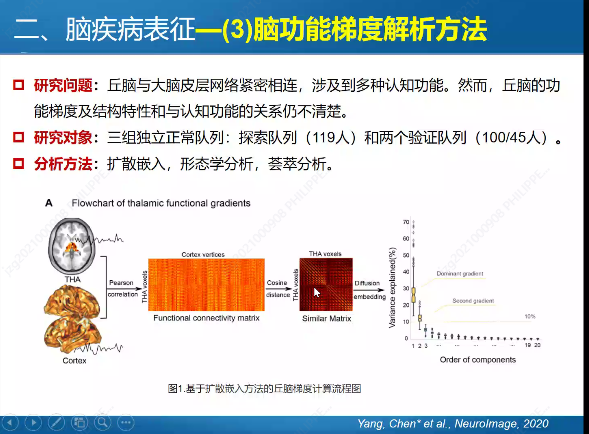
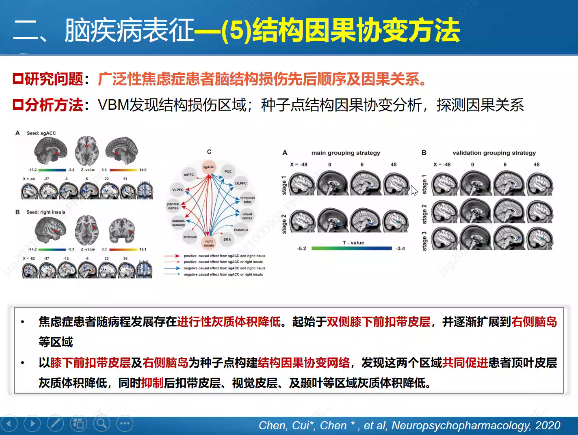
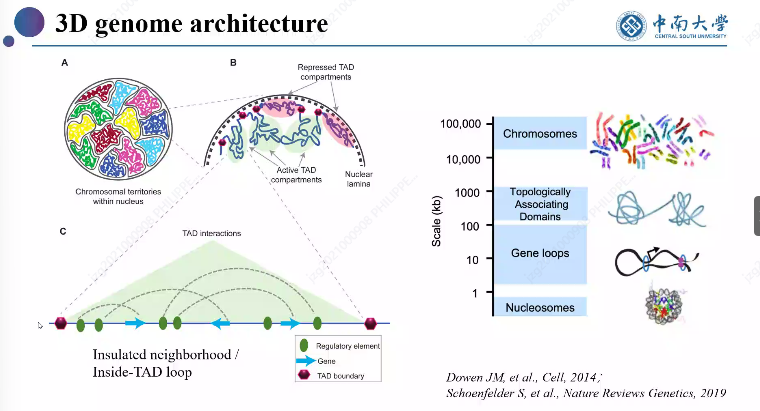
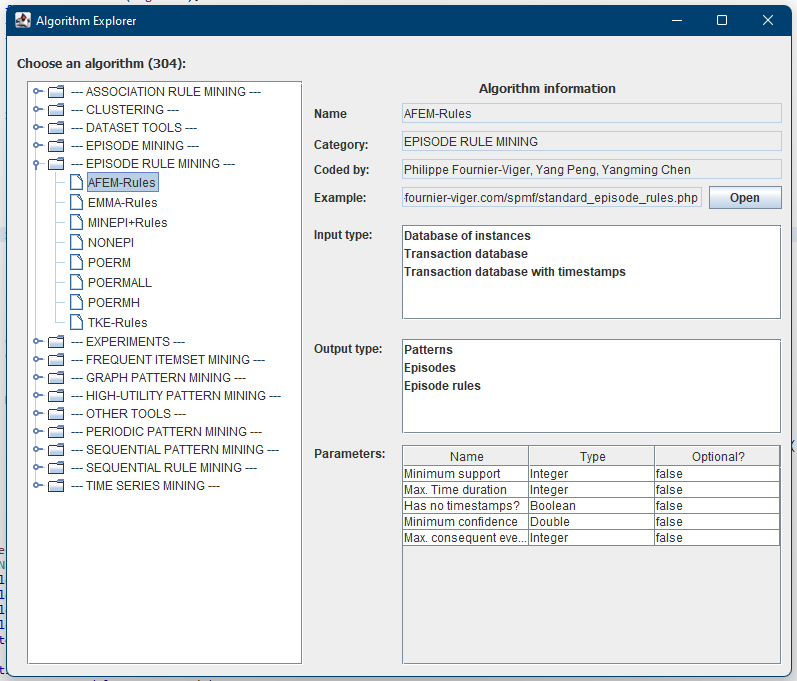
In previous blog posts, I have explained how to call SPMF as an external program from Python and how to call SPMF from C#. Today, I will explain how to call SPMF from an R program.
Requirements
Since SPMF is implemented in Java, the first requirement is to make sure that Java is installed on your computer. And of course, you need to also install R on your computer to run R programs.
Second, you should download the spmf.jar file from the SPMF website.
Third, you should make sure that your Java installation is correct. In particular, you should be able to execute the java command from the command line (terminal) of your computer because we will use the java command to call SPMF. If you type “java -version” in the command line of your computer, you should see the version of Java:

If you see this, then it is OK.
If you do not see this but instead get an error that java.exe is not found, it means that you have not installed Java, or that the PATH to Java is not setup properly on your computer so you cannot use it from the command line. If you are using the Windows operating System and you have installed Java, you need to make sure that java.exe is in the PATH environment variable. On Windows 11, you can fix this problem as follows: 1) Press WINDOWS + R, 2) Run the command “sysdm.cpl“, 3) Click the Advanced system settings tab. 4) Click Environment Variables. 5) In the section System Variables find the PATH environment variable and select it. 6) Click Edit. Add the path to the folder containing java.exe, which will be something like : C:\Program Files\Java\jdk-17.0.1\bin (depending on your version of Java and where you have installed it). 7) Click OK and close all windows. Then, you can open a new command prompt and try running “java -version” again to see if the problem is fixed. If you are using another version of Windows or the Linux operating system, you can find similar steps online about how to setup Java on your computer.
1) Launching the GUI of SPMF from R
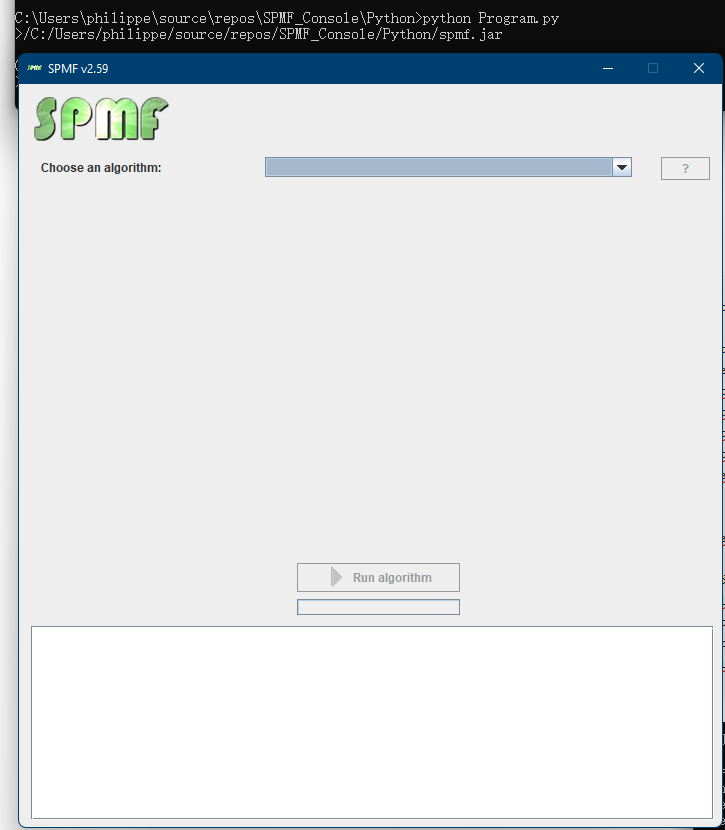
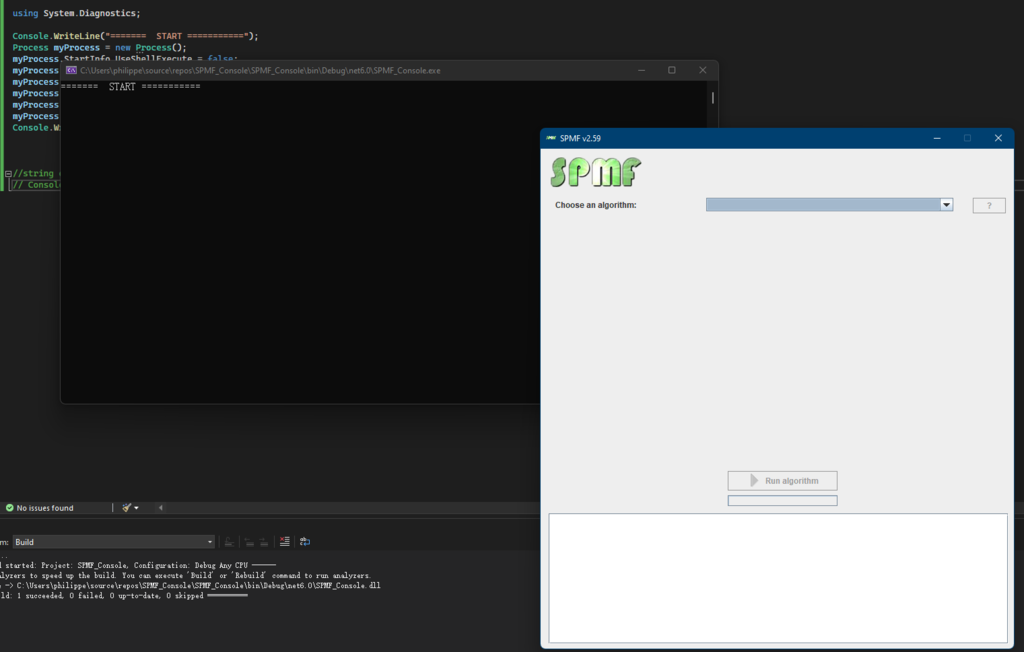
Now that I have explained the basic requirements, I will first show you how to launch the GUI of SPMF from R. For this, it is very simple. Here I give you the code of a simple R program that calls the Jar file of SPMF to launch the GUI of SPMF.
#Set the working directory to the folder containing spmf.jar
setwd("C:\\Users\\philippe\\Desktop\\")
#Call SPMF as an external program
system("java -jar C:\\Users\\philippe\\Desktop\\spmf.jar")
What this program does? It basically just runs the command
java -jar spmf.jar
By running this program, SPMF is successfully launched:
2) Executing an algorithm from SPMF from a R program
Next, I will explain something more useful, that is how to run an algorithm from SPMF from an R program? We will modify the above program to do this. Let’s say that we want to run the Apriori algorithm on an input file called contextPasquier99.txt (this file is included with SPMF and can be downloaded here).
To do this, we need to first check the documentation of SPMF to see how to run the Apriori algorithm from the command line. The documentation of SPMF is here. How to run Apriori is explained in this page of the documentation. We find that we can use this command
java -jar spmf.jar run Apriori contextPasquier99.txt output.txt 40%
to run Apriori on the file contextPasquier99.txt with the parameter minsup = 40% and to save the result to a file output.txt.
To do this from R, we can write a simple R program like this:
#Set the working directory to the folder containing spmf.jar and the file contextPasquier99.txt
setwd("C:\\Users\\philippe\\Desktop\\")
#Call SPMF as an external program to run the Apriori algorithm
system("java -jar spmf.jar run Apriori contextPasquier99.txt output.txt 40%")
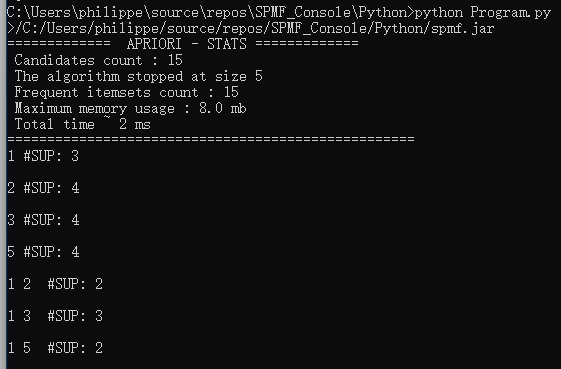
Then, it will produce the file output.txt as result in the workind directory:
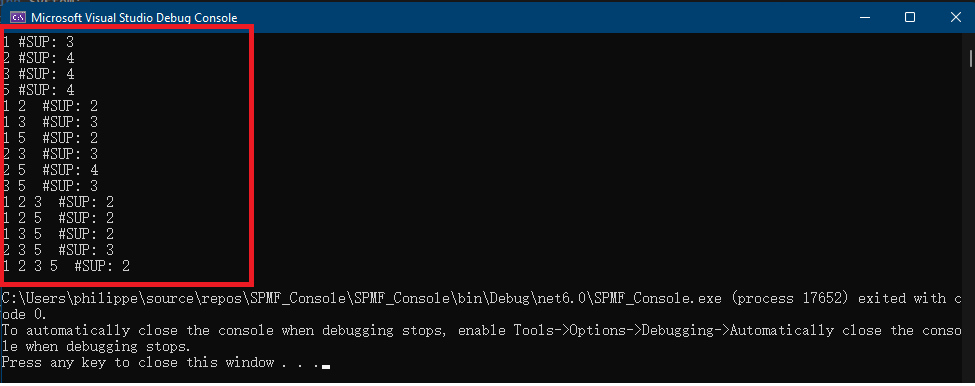
If we open the file “output.txt”, we can see the content:
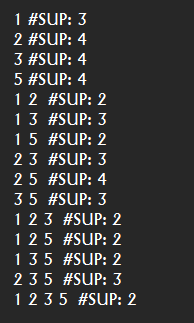
Each line of this file is a frequent itemset found by the Apriori algorithm. To understand the input and output file format of Apriori, you can see the documentation of the Apriori algorithm.
If you want to call other algorithms that are offered in SPMF besides Apriori, you can lookup the algorithm that you want to call in the SPMF documentation. An example is provided for each algorithm in the SPMF documentation and explanation of how to run it.
3) Executing an algorithm from SPMF from a R program and then reading the output file
Now, I will explain how to read the output file produce by SPMF from an R program. When running an algorithm of SPMF such as in the previous example, the output is generally a text file. We can easily read an output file from R to obtain the content.
For instance, I modified the previous R program to read the content of the file “output.txt” that is produced by SPMF to show its content in the console. The new R program is below:
#Set the working directory to the folder containing spmf.jar and the file contextPasquier99.txt setwd("C:\\Users\\philippe\\Desktop\\")
#Call SPMF as an external program to run the Apriori algorithm
system("java -jar spmf.jar run Apriori contextPasquier99.txt output.txt 40%")
# Read the output file line by line and print to the console
myCon = file(description = "output.txt", open="r", blocking = TRUE)
repeat{
pl = readLines(myCon, n = 1) # Read one line from the connection.
if(identical(pl, character(0))){break} # If the line is empty, exit.
print(pl) # Otherwise, print and repeat next iteration.
}
close(myCon)
rm(myCon)
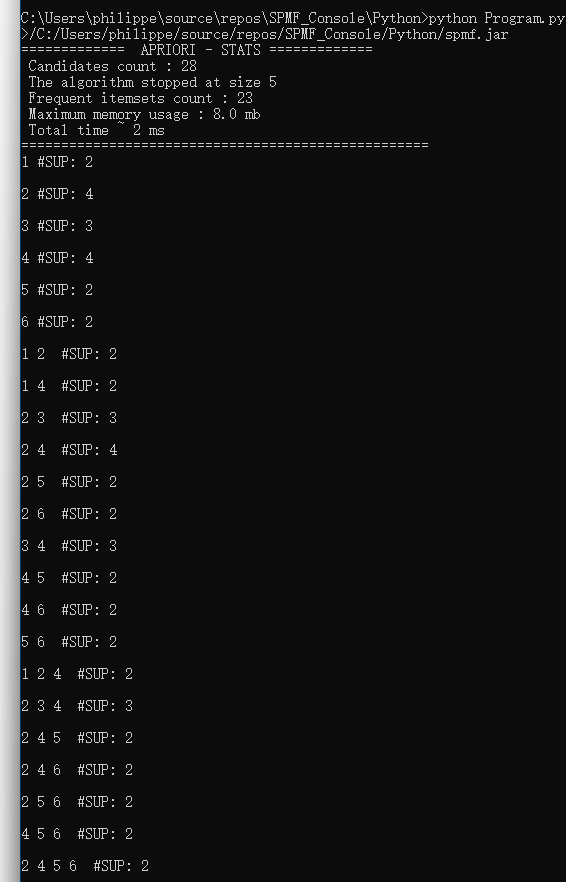
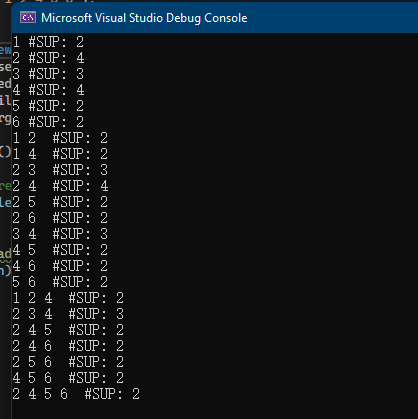
If we execute this R program, it will first call the Apriori algorithm from SPMF. Then, the R program will read the content of the output file output.txt line by line and display the content in the console:

We could further modify this program to do something more meaningful with the content of the output file such as reading the content in R data frames to do further processing. But at least, I wanted to show you the basic idea of how to read an output file from SPMF from an R program.
3) Writing an input file for SPMF from a R program, and then running an algorithm from SPMF
Lastly, you can also write the input file that is given to SPMF from a R program by using code to write a text file.
For example, I will modify the example above to write a new text file called “input.txt” that will contain the following data:
1 2 3 4
2 3 4
2 3 4 5 6
1 2 4 5 6
and then I will call SPMF to execute the Apriori algorithm on that file. Then, the program will read the output file “output.txt” from R. Here is the code:
#Set the working directory to the folder containing spmf.jar and the file contextPasquier99.txt setwd("C:\\Users\\philippe\\Desktop\\")
# Write an input file for Apriori
file.create("input.txt")
sink("input.txt")
cat("1 2 3 4\r\n")
cat("2 3 4\r\n")
cat("2 3 4 5 6\r\n")
cat("1 2 4 5 6")
sink()
#Call SPMF as an external program to run the Apriori algorithm
system("java -jar spmf.jar run Apriori input.txt output.txt 40%")
# Read the output file line by line and print to the console
myCon = file(description = "output.txt", open="r", blocking = TRUE)
repeat{
pl = readLines(myCon, n = 1) # Read one line from the connection.
if(identical(pl, character(0))){break} # If the line is empty, exit.
print(pl) # Otherwise, print and repeat next iteration.
}
close(myCon)
rm(myCon)
After running this program, the file “input.txt” is successfully created:
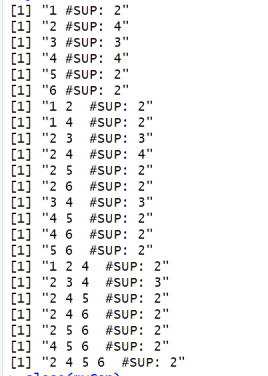
And the content of the output file output.txt is shown in the console:
Conclusion
In this blog post, I have shown the basic idea of how to call SPMF from R by calling SPMF as an external program. It is quite simple. It just require to know how to read/write files in R.
Hope that this has been interesting.
==
Philippe Fournier-Viger is a full professor and the founder of the open-source data mining software SPMF, offering more than 110 data mining algorithms. If you like this blog, you can tweet about it and/or subscribe to my twitter account @philfv to get notified about new posts.