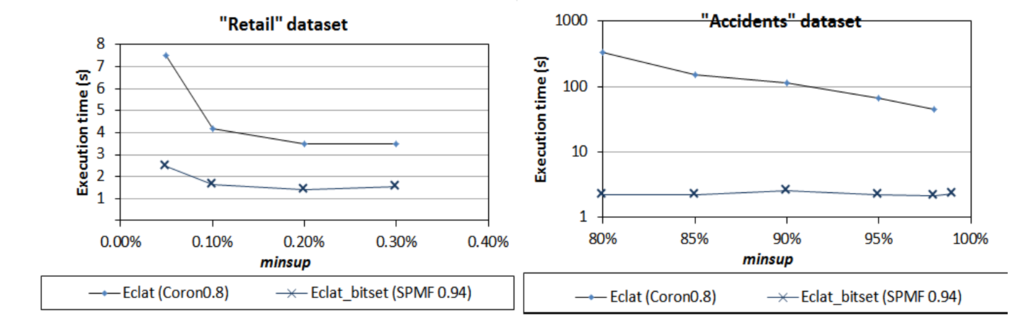
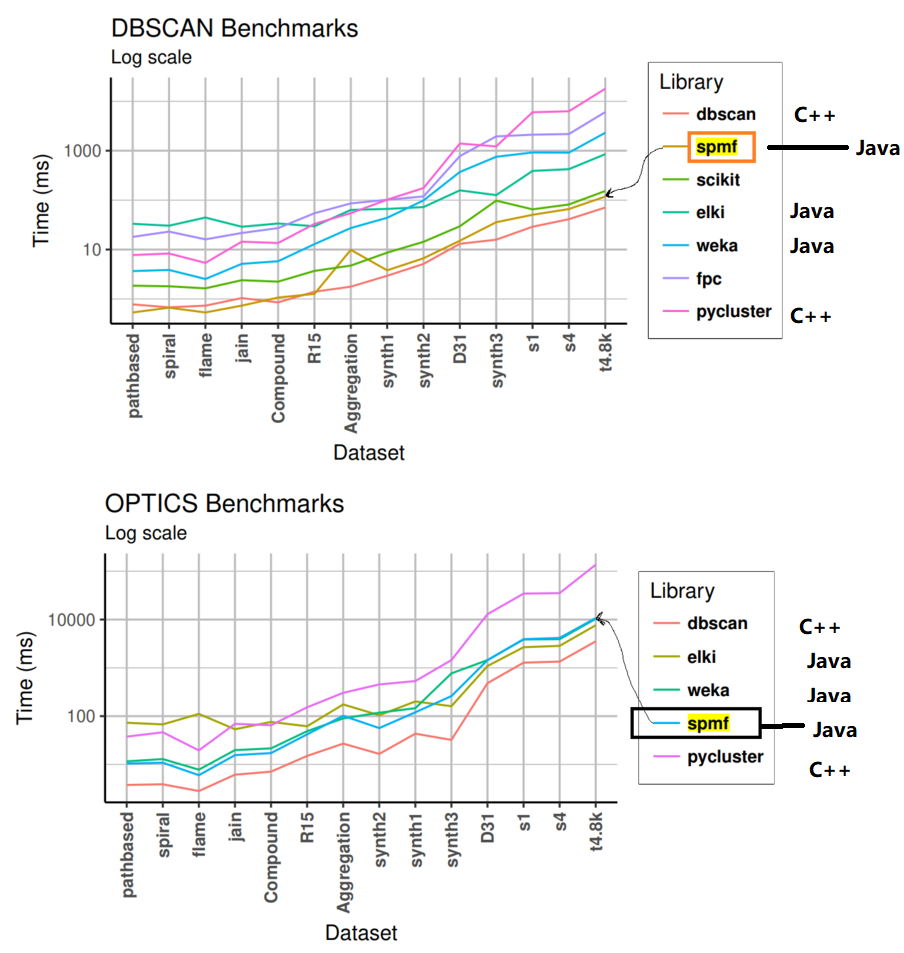
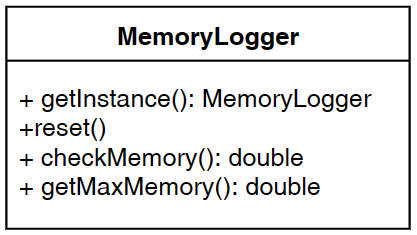
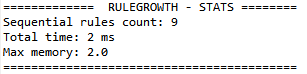
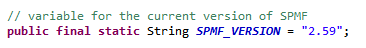
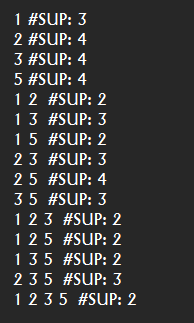
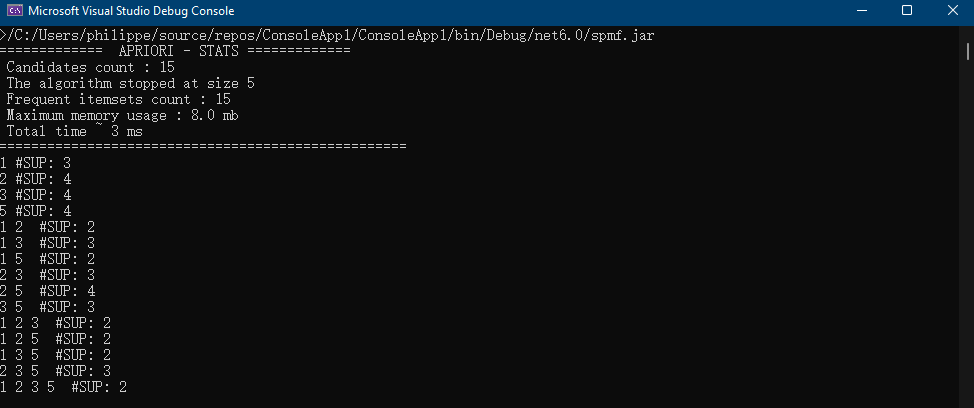
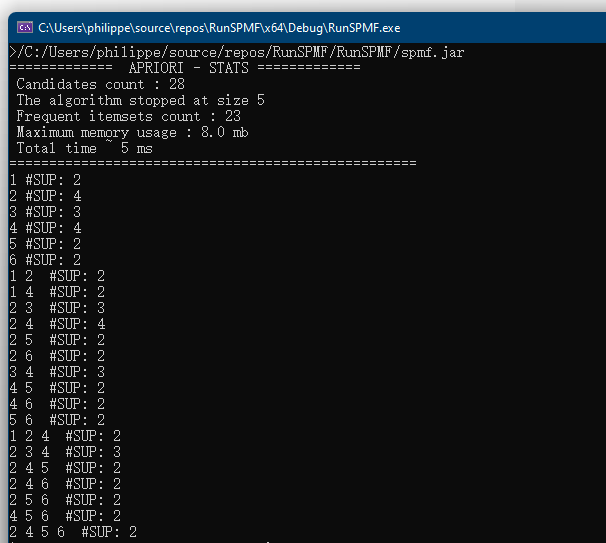
This is the fifth post of a series of blog posts about the architecture of the SPMF data mining library. Today, I will specifically talk about the graphical user interface of SPMF, its main components and how it interacts with other components from SPMF.
An overview of the architecture of SPMF
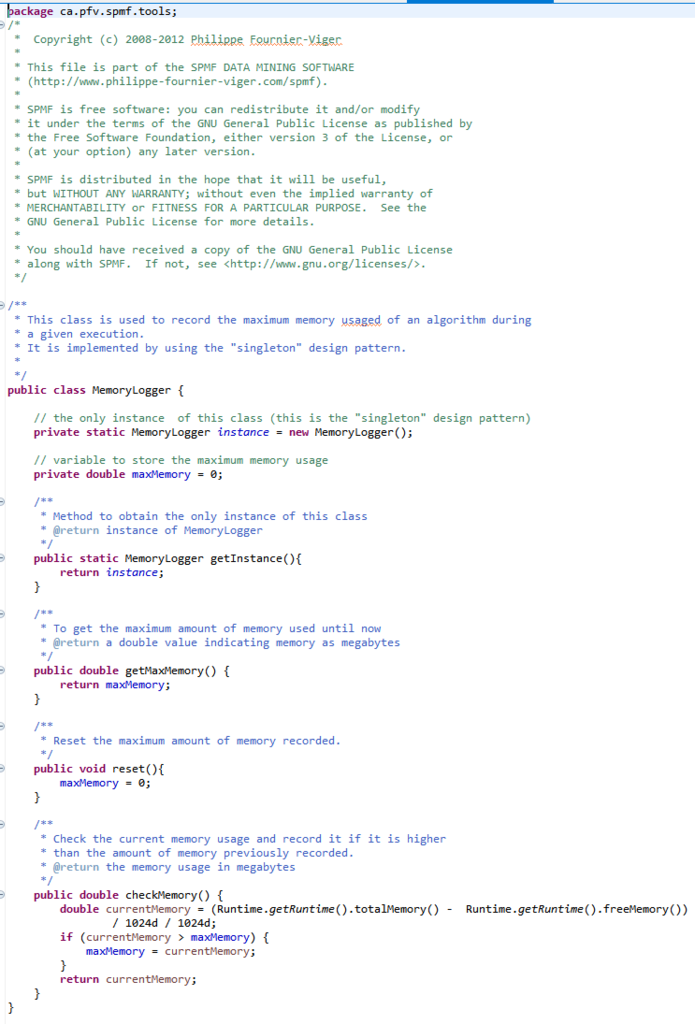
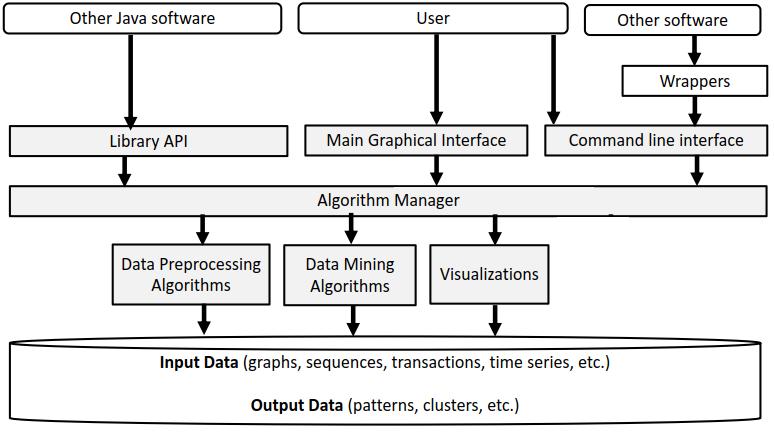
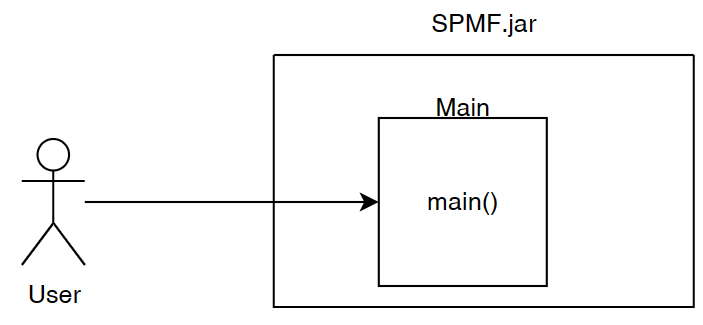
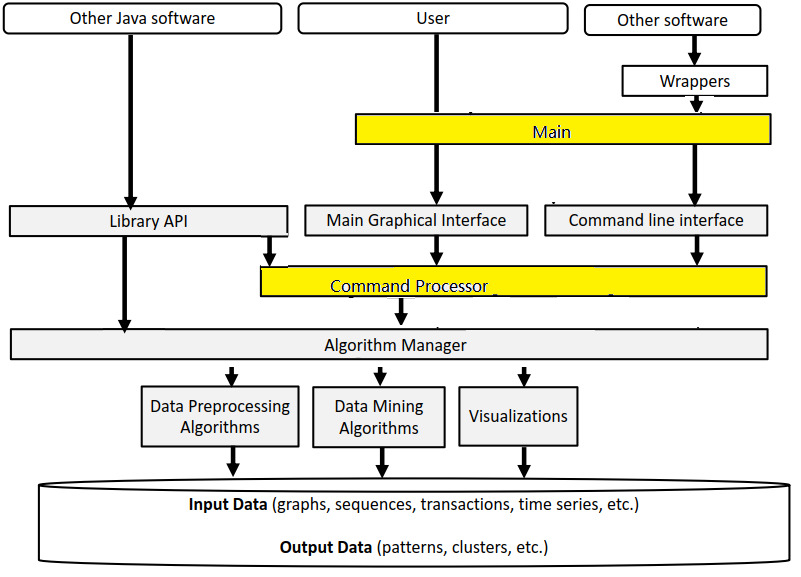
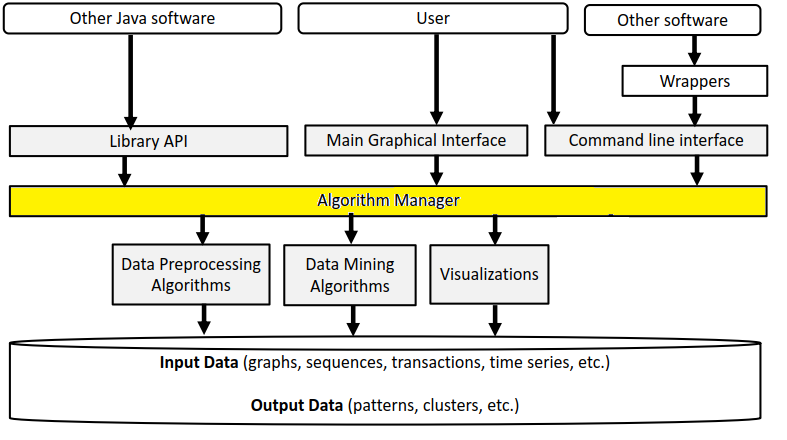
Before, I start talking about the graphical user interface of SPMF, let’s look at the overall architecture of SPMF:
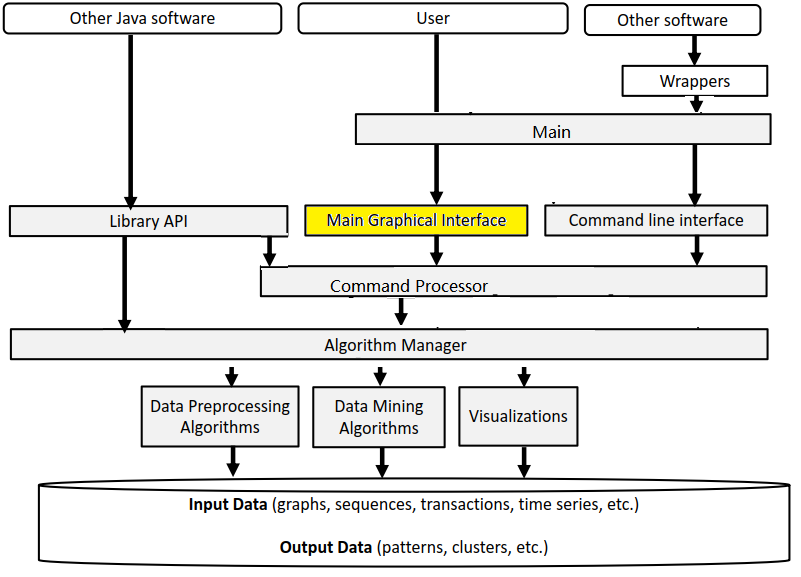
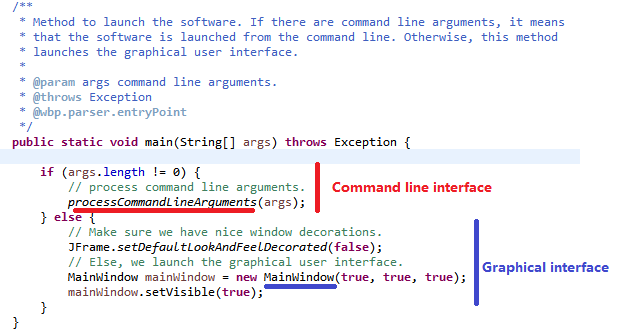
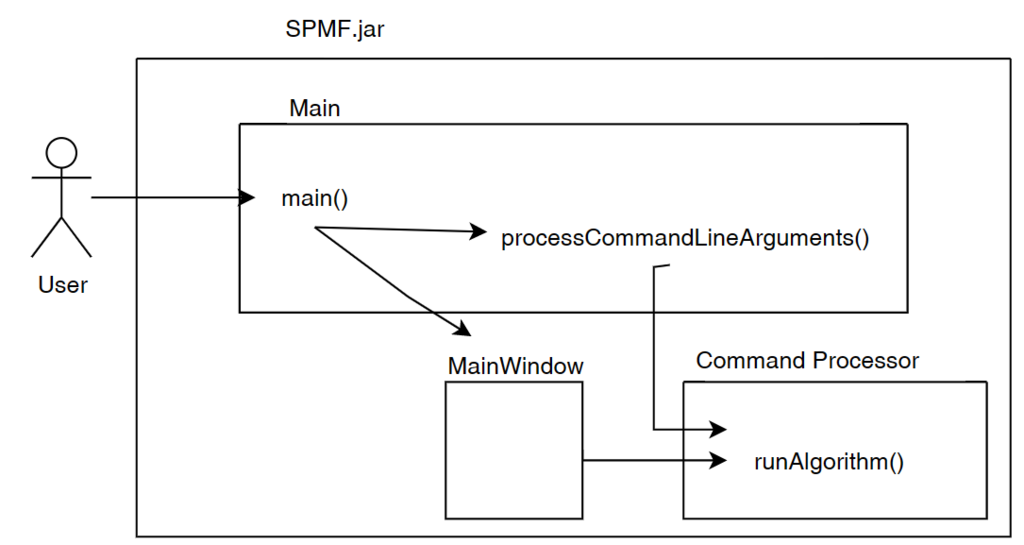
SPMF is implemented using the Java language. It can be used in three ways: (1) using a simple graphical user interface, (2) using its command line interface or (3) by integrating it into other software directly as a Java library or using some wrappers. When running the SPMF software as a standalone program, the Main class is the entry point of the software, which then calls the graphical interface or the command line interface. The user can use these interfaces to select some algorithm to be run with some parameters. Running an algorithm is then done by calling a module call the Command Processor, which obtains information about the available algorithms from another module called the Algorithm Manager.
Now for today’s topic, I will talk in more details about the graphical interface.
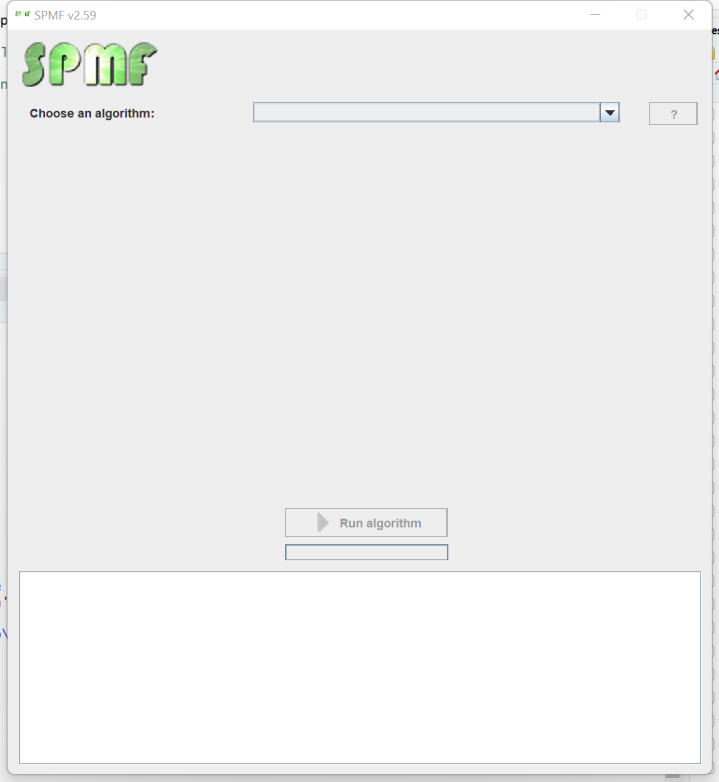
The Graphical Interface
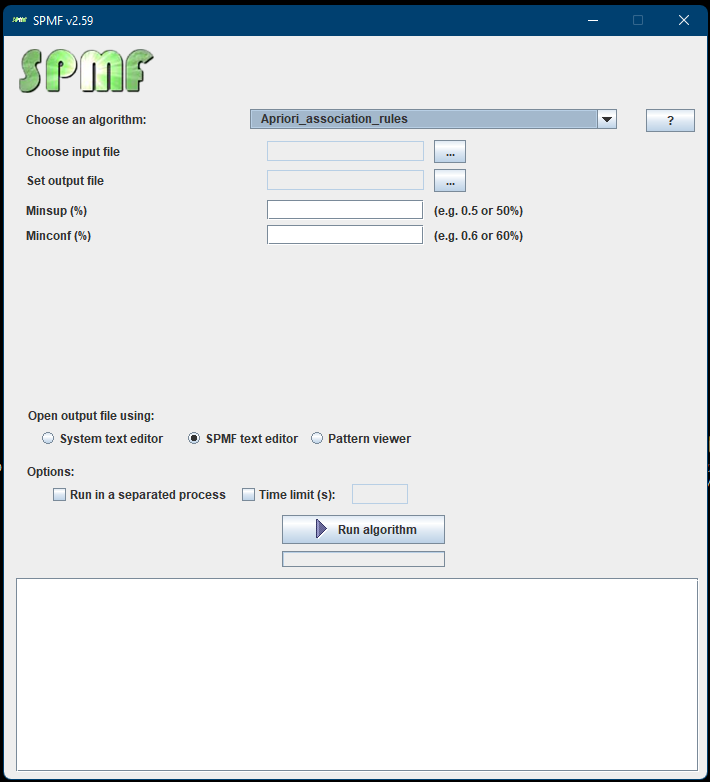
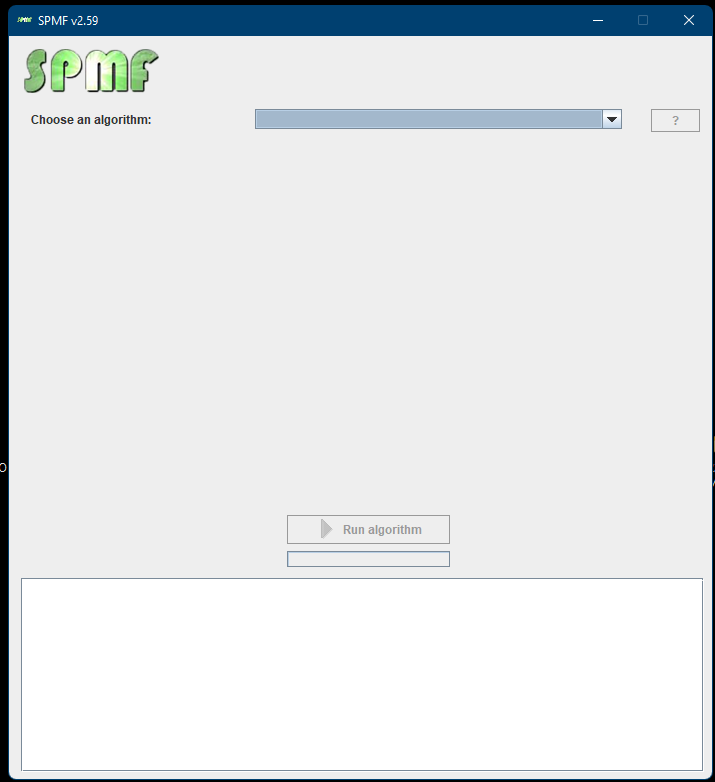
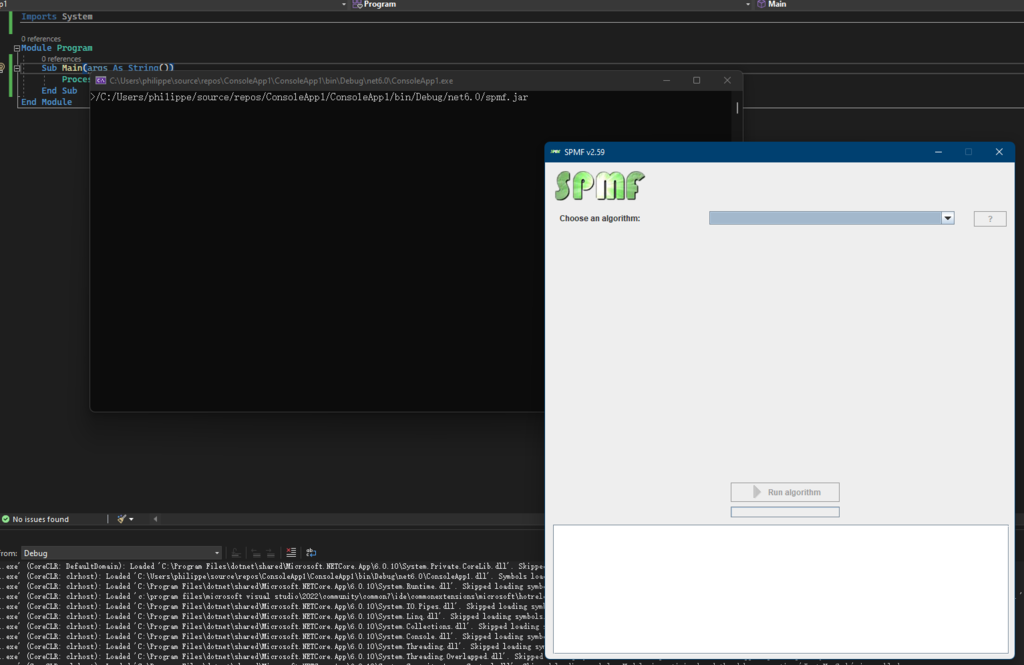
The main class of the Graphical Interface in SPMF is called MainWindow and is located in the package ca.pfv.spmf.gui. The class MainWindow contains the code for the main Window of SPMF, which looks like that:
This Window is implemented using the Java Swing library for building a graphical user interface using Java.
In the package ca.pfv.spmf.gui, there are also several other classes that are used for other aspects of the user interface, such as for displaying visualizations of datasets and results from data mining algorithms. I will talk more about this later.
What are the relationships between the Graphical Interface and other components of SPMF?
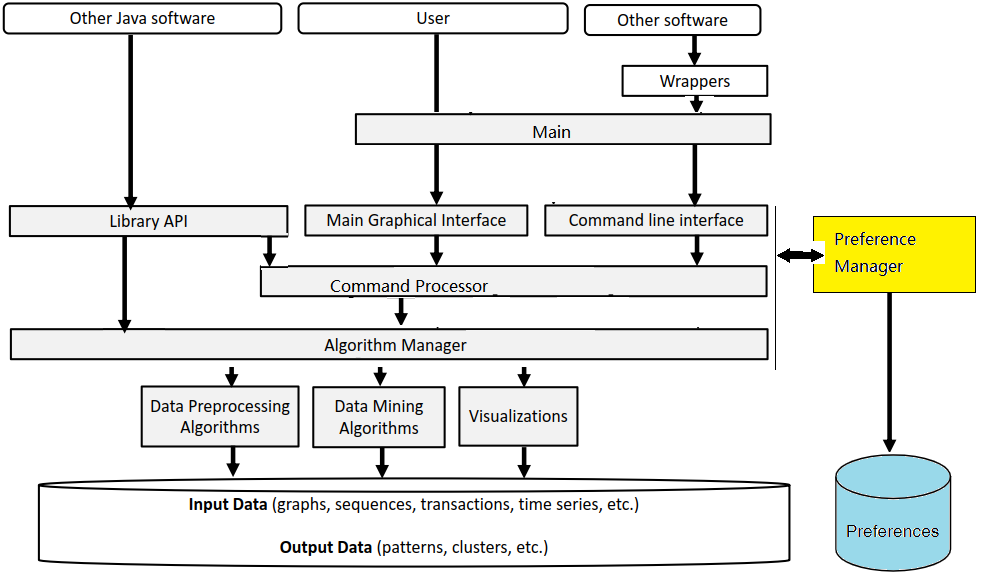
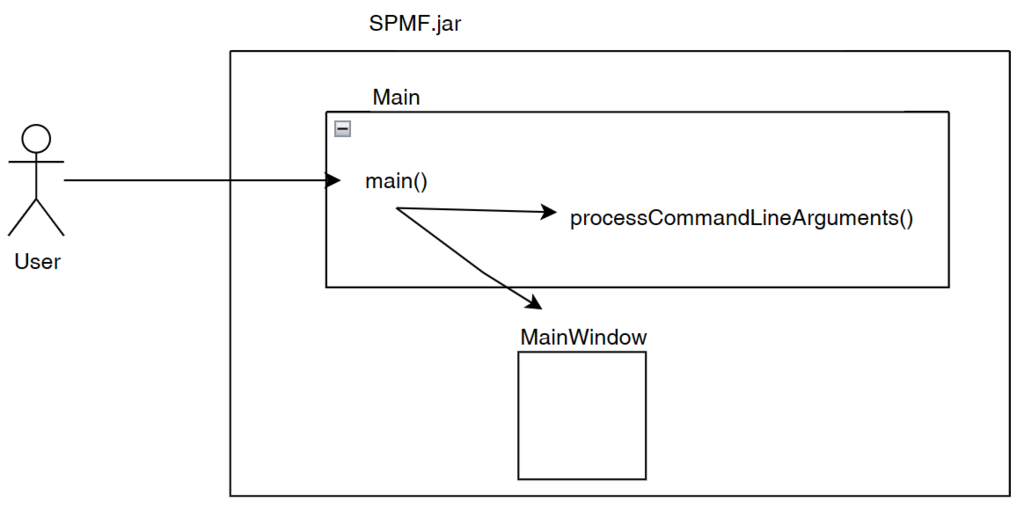
I would like now to explain how the graphical interface interacts with other components of SPMF. Here is a small illustration:
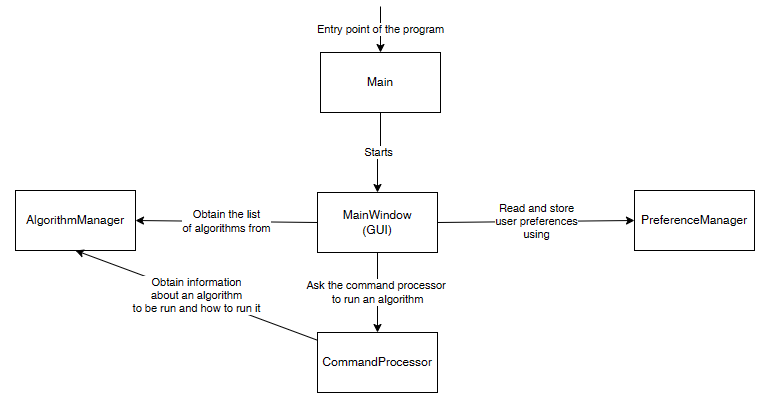
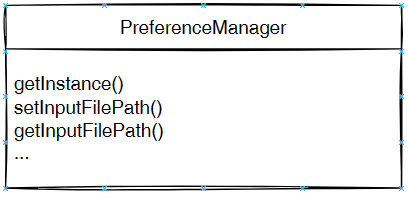
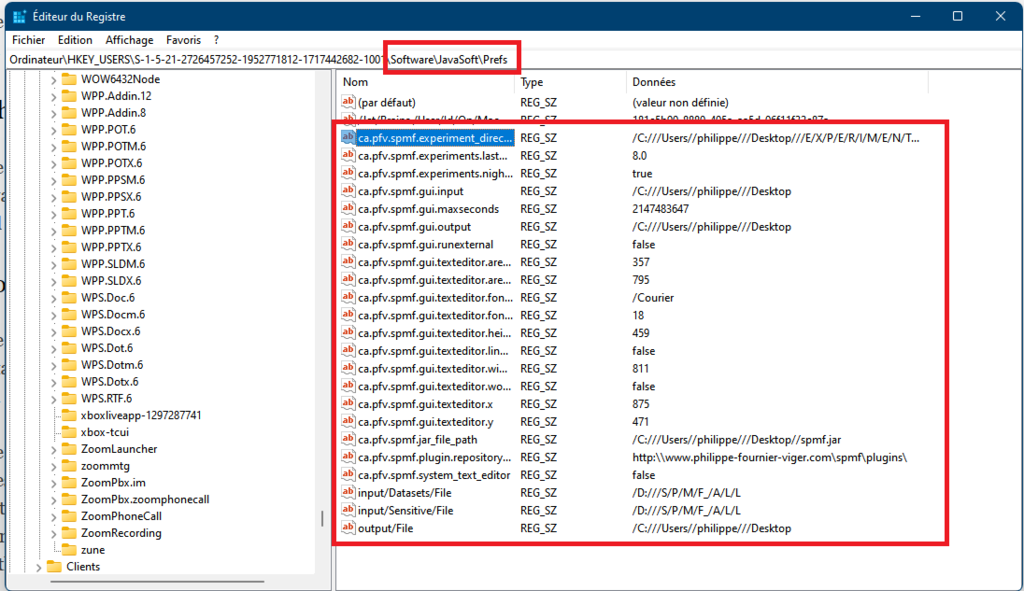
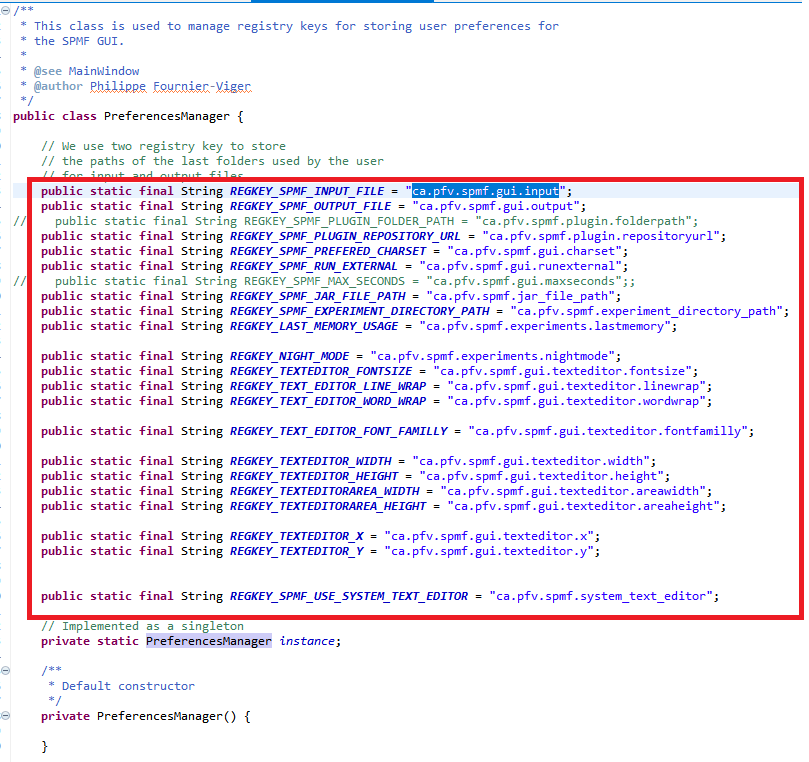
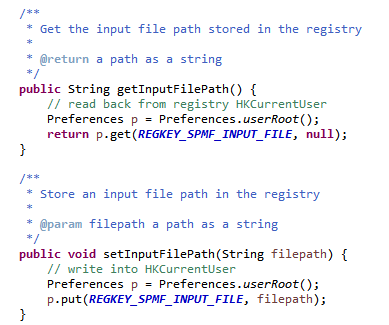
As said, previously, when running SPMF, the Main class is starting the graphical interface by creating an instance of the MainWindow class. The MainWindow and graphical user interface of SPMF in general reads and write user preferences through another module called the PreferenceManager. The type of user preferences that are stored by SPMF are for example, the last directory that the user has used for reading an input file using SPMF and the preferences of using the text editor of SPMF or the system’s text editor to open output files. The PreferenceManager stores these preferences permanently, using for example the Window Registry if SPMF is run using the Windows operating system.
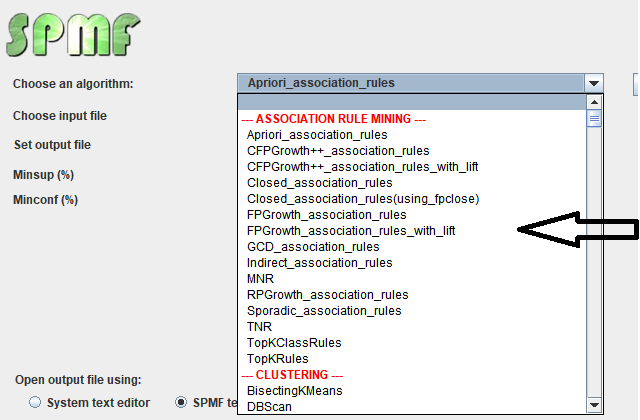
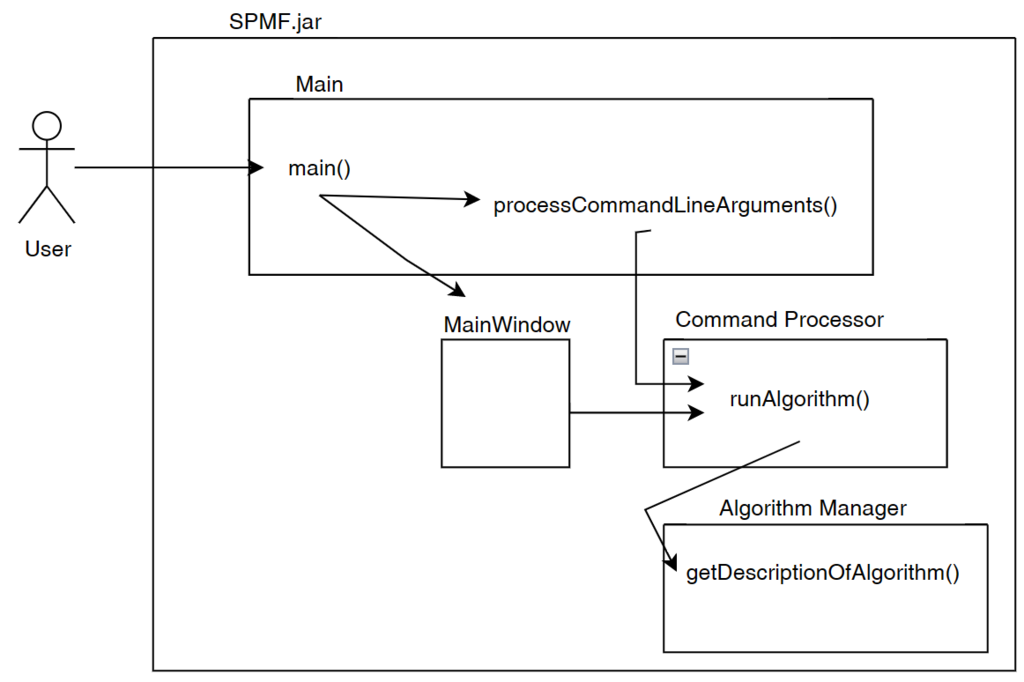
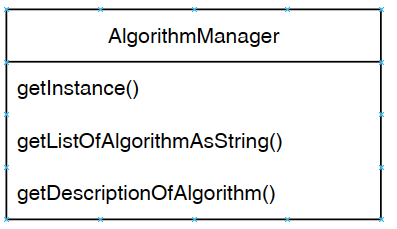
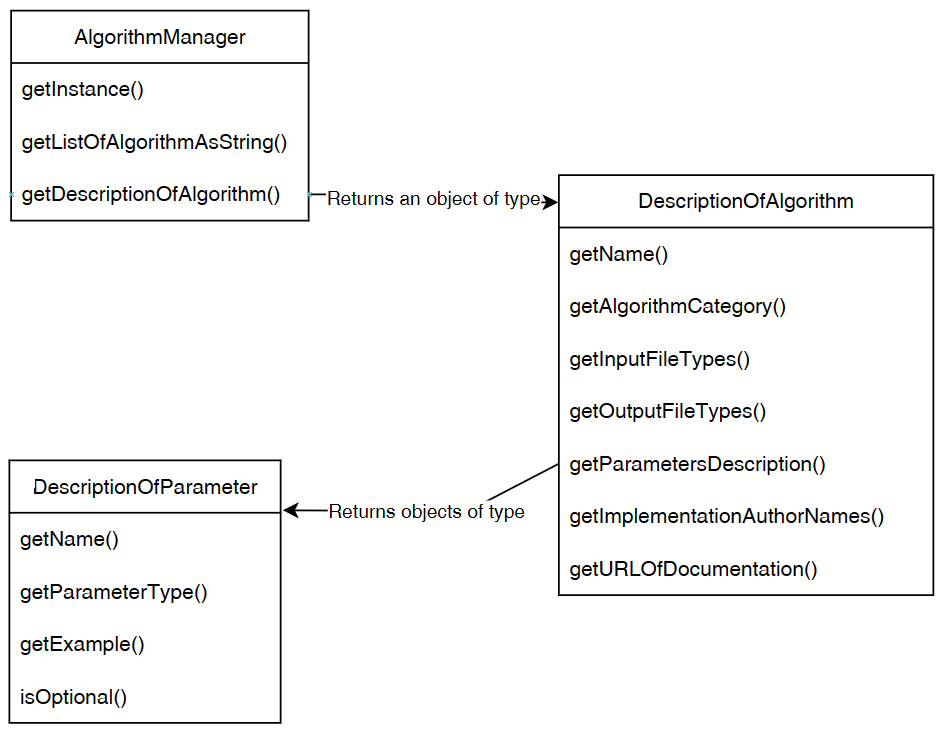
Besides that, the graphical user interface is also interacting with the the Algorithm Manager module of SPMF to obtain the list of available algorithms and information about them such as the required parameters. This allows the graphical interface to display the list of algorithms to the user:
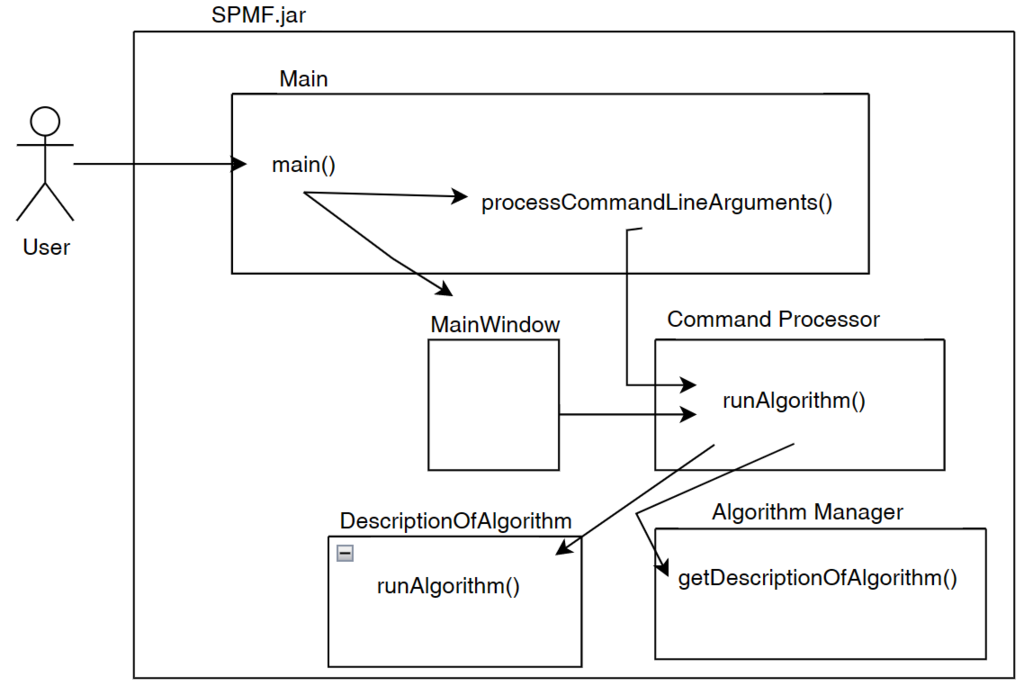
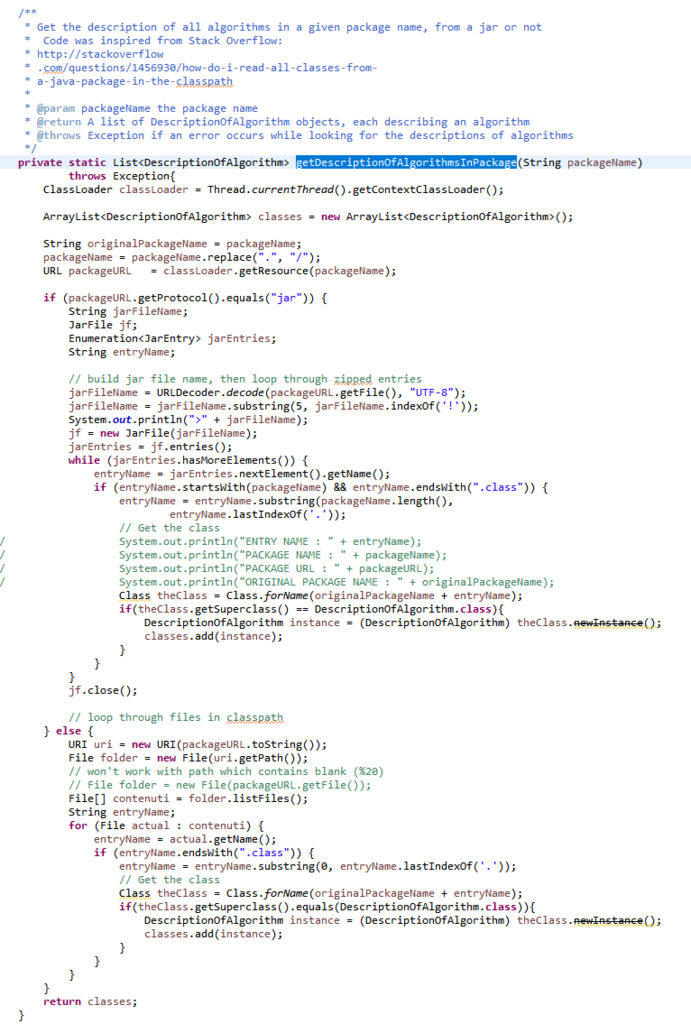
Now, after the user selects and algorithm to run it, the action of the running the algorithm is done by calling the Command Processor module. That module takes care of verifying that the parameters provided by the user are appropriate for the algorithm that has been selected, obtain information about how to run the algorithm from the Algorithm Manager, and then run the algorithm. Besides, the Command Processor can also automatically convert some special file types before calling an algorithm. For example, the Command Processor can automatically transform ARFF and TEXT files to the SPMF format so that algorithms from SPMF can also be run on ARFF files and TEXT documents with a .text extension.
What are the main classes of the Graphical Interface?
It would take a long time to describe all the classes that make up the graphical interface of SPMF. But mainly, all the classes are located in the package ca.pfv.spmf.gui and there is at least one class for each window used by SPMF. Here is an overview:
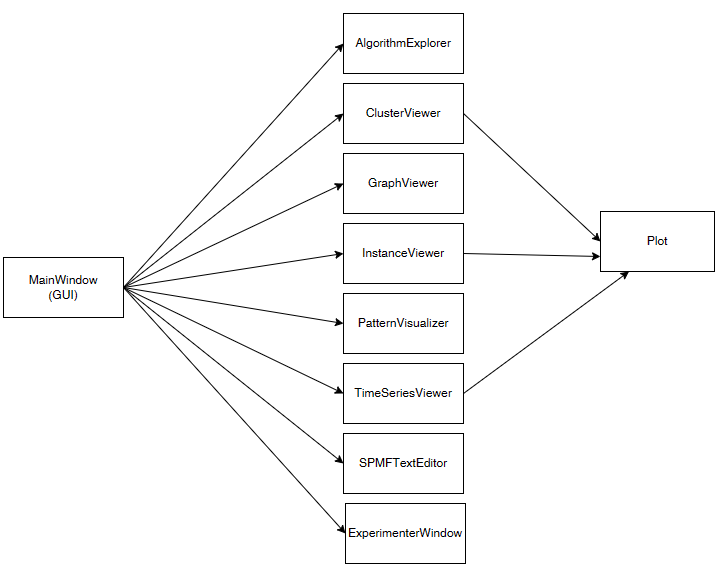
In the middle, you can see the main classes for the different windows from the graphical interface:
- AlgorithmExplorer: This windows lets the user browse the different algorithms offered in SPMF and get detailed information about them.
- ClusterViewer: This windows is used to display clusters found by clustering algorithms such as K-Means.
- GraphViewer: This windows is used to display frequent subgraphs found by graph mining algorithms or to display graph datasets
- InstanceViewer: This windows is used to display the content of input data files for clustering. These files are vectors of numbers
- PatternsVisualizer:This windows can display the patterns found by a data mining algorithm that is applied by the user, using a table view. It offers various functions such as to search, sort and filter patterns.
- TimeSeriesViewer: This windows is used for displaying time series data visually.
- SPMFTextEditor: This is a simple text editor that is provided with SPMF. It can be used to display the output files produced by the algorithms.
- ExperimenterWindow: This is a window to run performance experiments to test some algorithms. For example, it allows to automatically compare the performance of multiple algorithms on a dataset while varying a parameter.
- AboutWindow: This window presents general information about SPMF.
Several windows such as the InstanceViewer, ClusterViewer and TimeSeriesViewer are displaying plots. This is done using the Plot class from the ca.pfv.spmf.gui.plot package.
How the graphical user interface was implemented?
A large part of the user interface of SPMF was coded by hand in Java. But there also exists some tools that can help in building a user interface more quickly when developing Java programs. For programming SPMF, I mostly used the Eclipse IDE (Integrated Development Environment), and there exists a plugin called the WindowBuilder Editor, which allows to create user interfaces visually. This can save quite some time for user interface programming. For example, here is a screenshot of that plugin for designing the TimeSeriesViewer of SPMF:
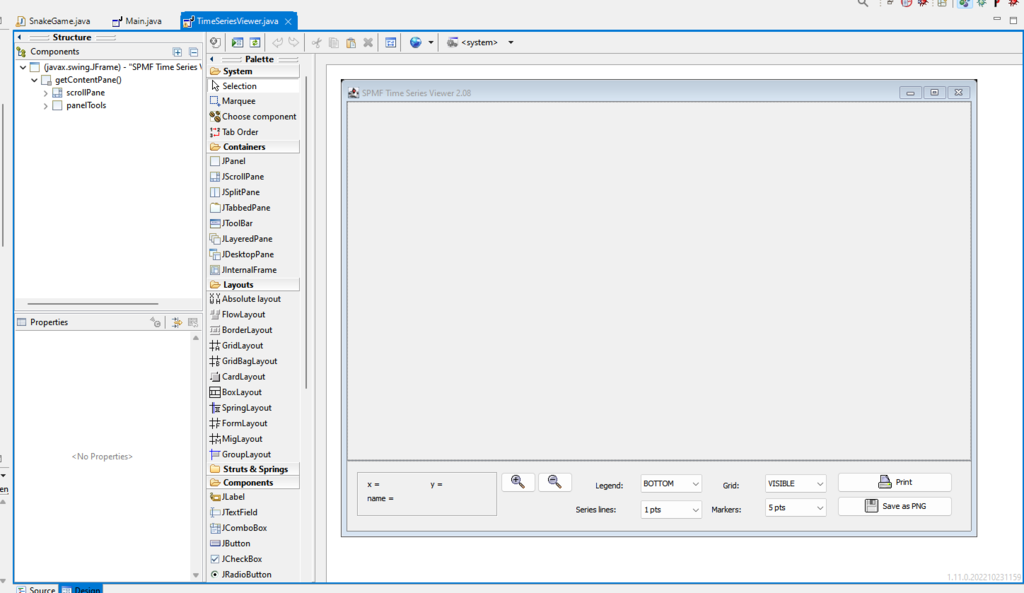
Using the WindowBuilder Editor, I can directly place different components using the mouse such as buttons, text fields, and combo boxes. Then, after that, I can connect these components to events such as mouse clicks and write Java code for handling the events.
Conclusion
Today, I have given an overview about how the user interface of SPMF is designed, its main components and how it interacts with other components from the SPMF software. Hope it has been interesting!
==
Philippe Fournier-Viger is a full professor and the founder of the open-source data mining software SPMF, offering more than 250 data mining algorithms.