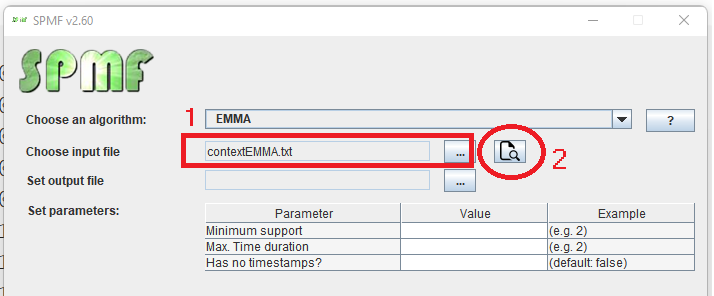
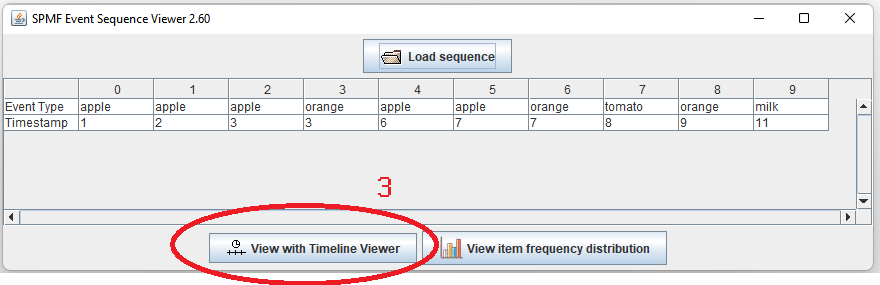
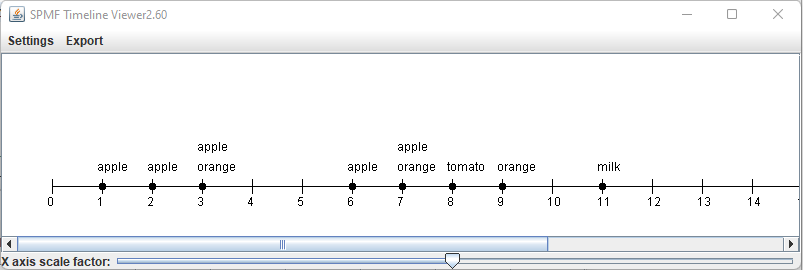
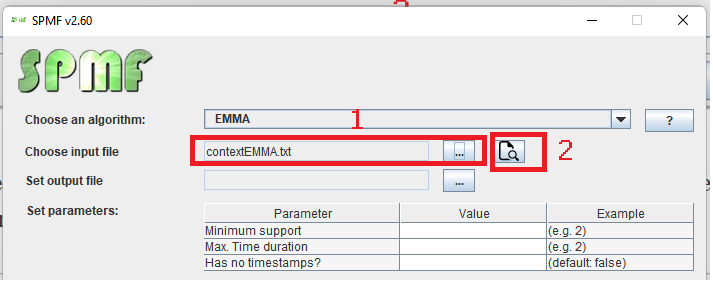
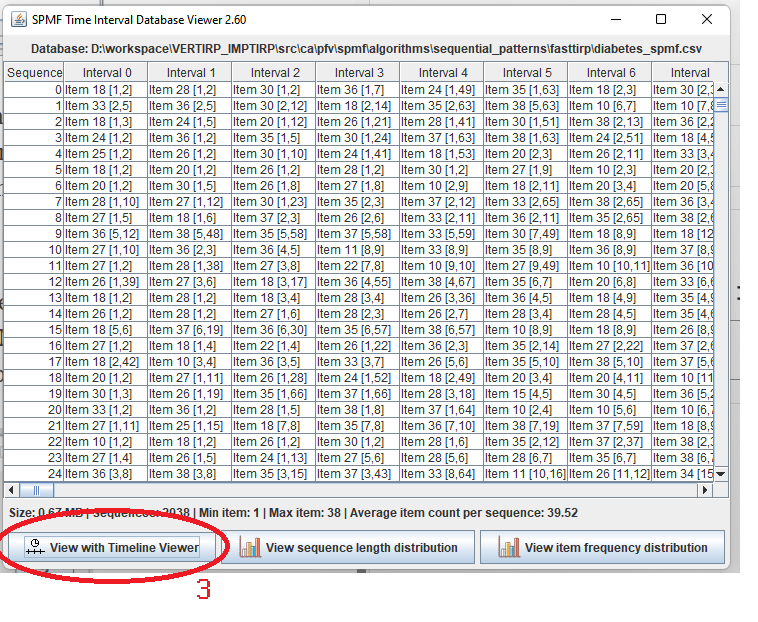
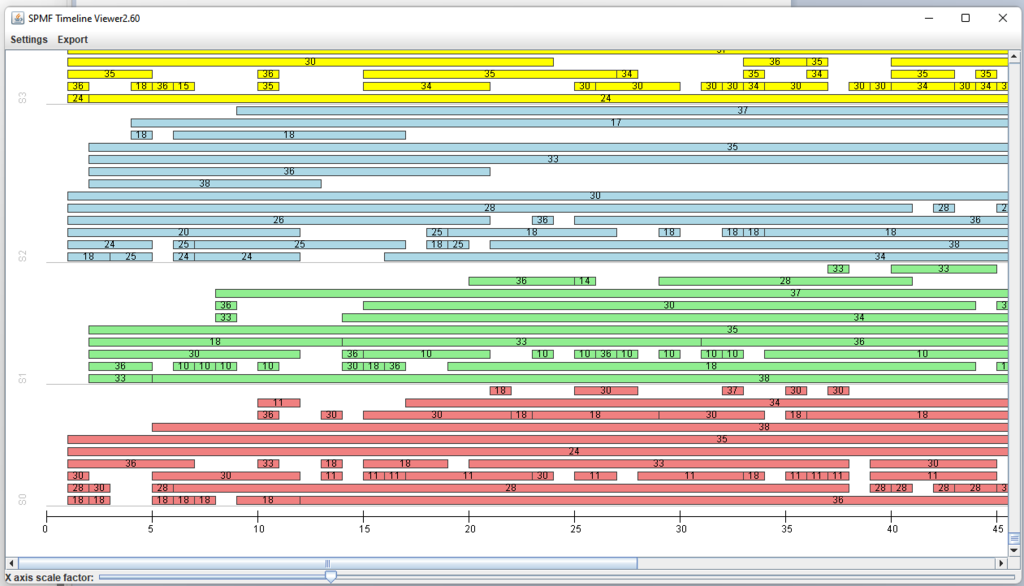
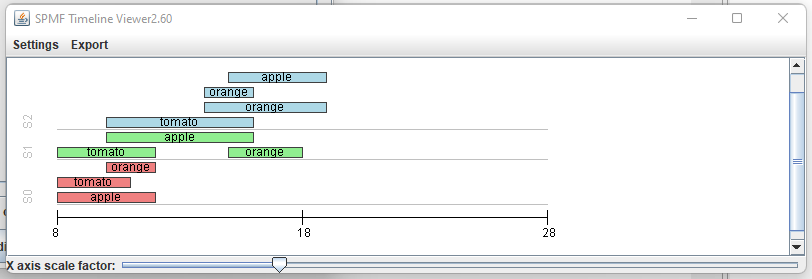
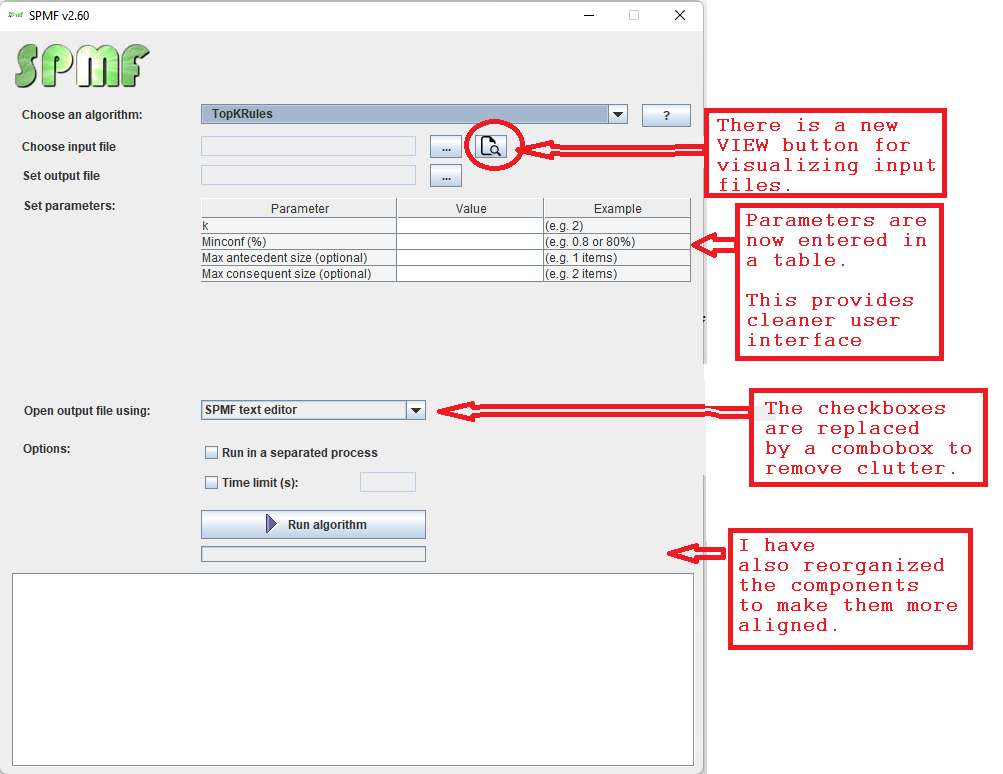
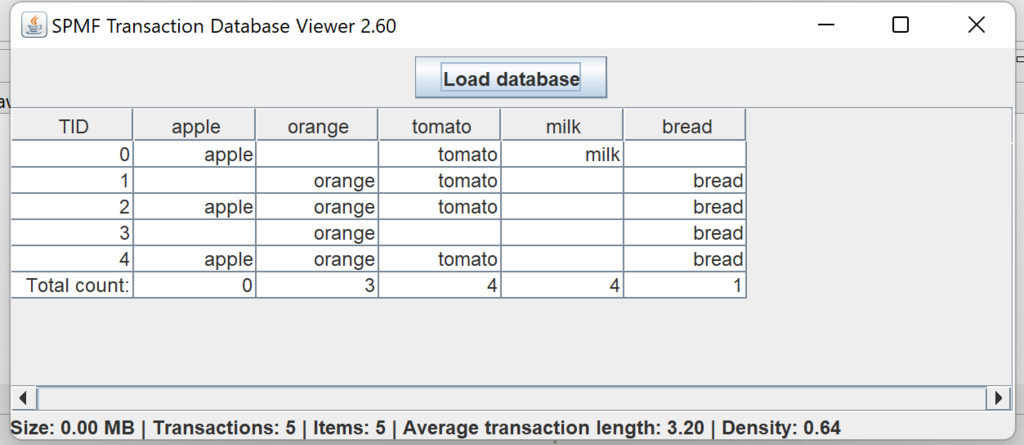
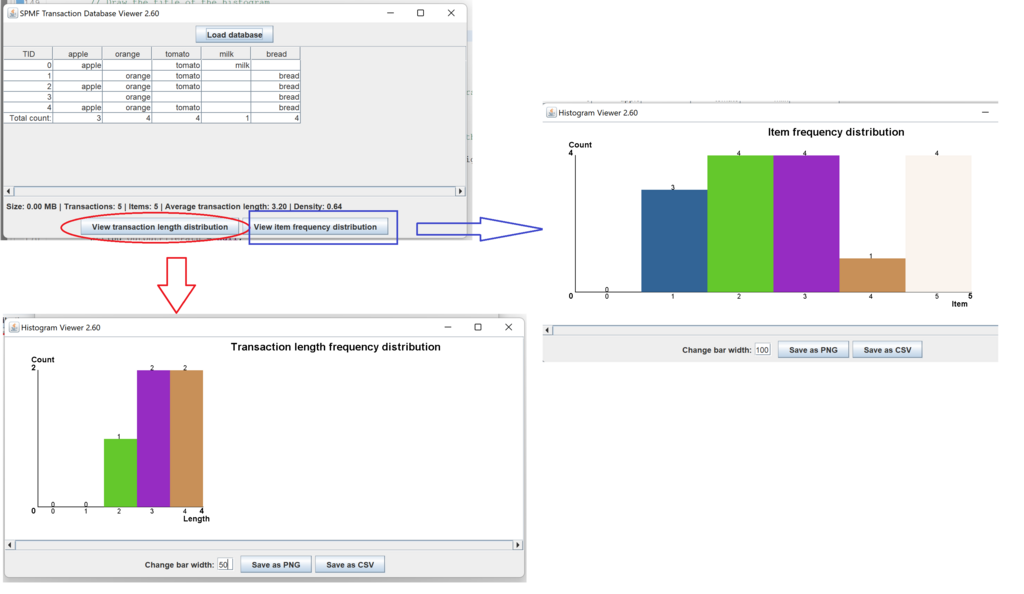
This is a short message today to announce that the new version of SPMF 2.60 is finally released!
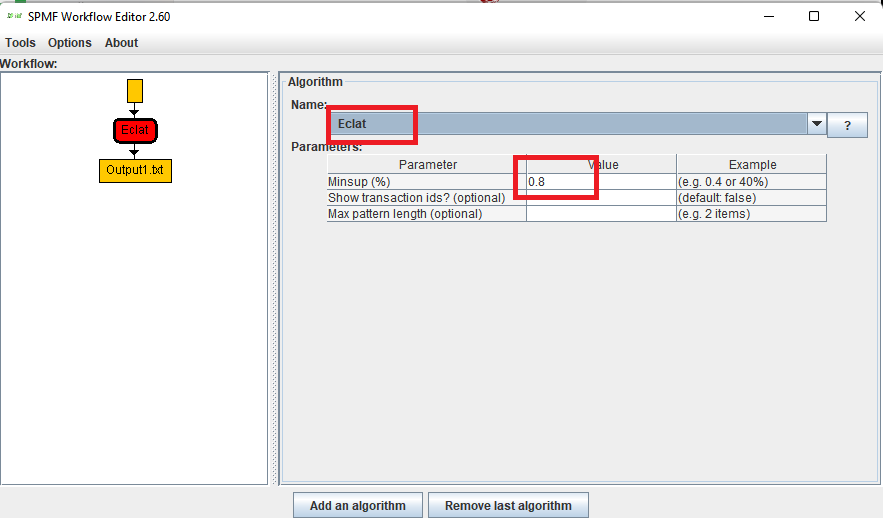
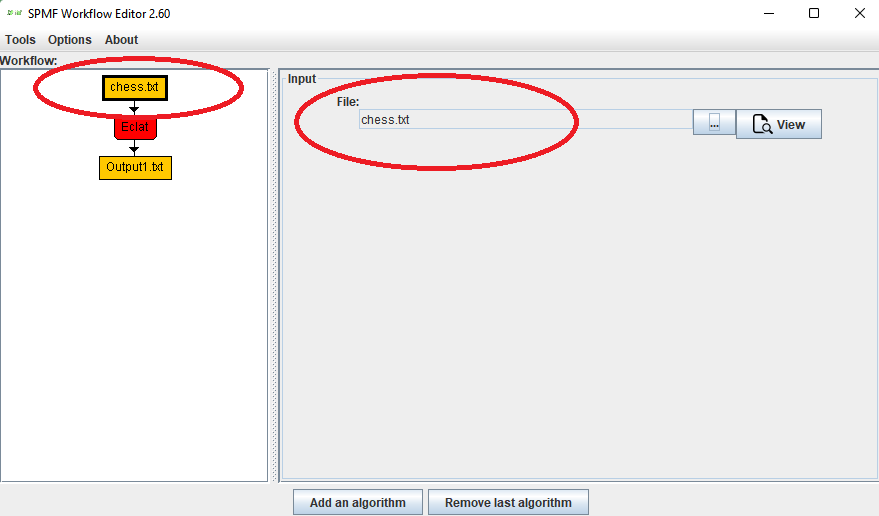
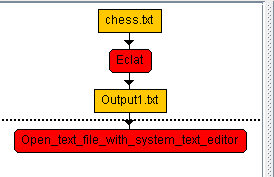
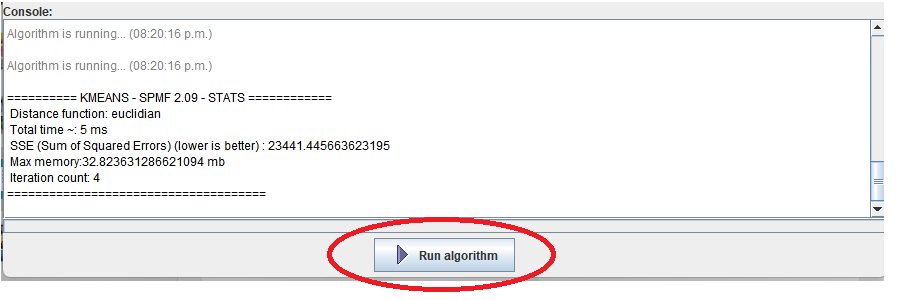
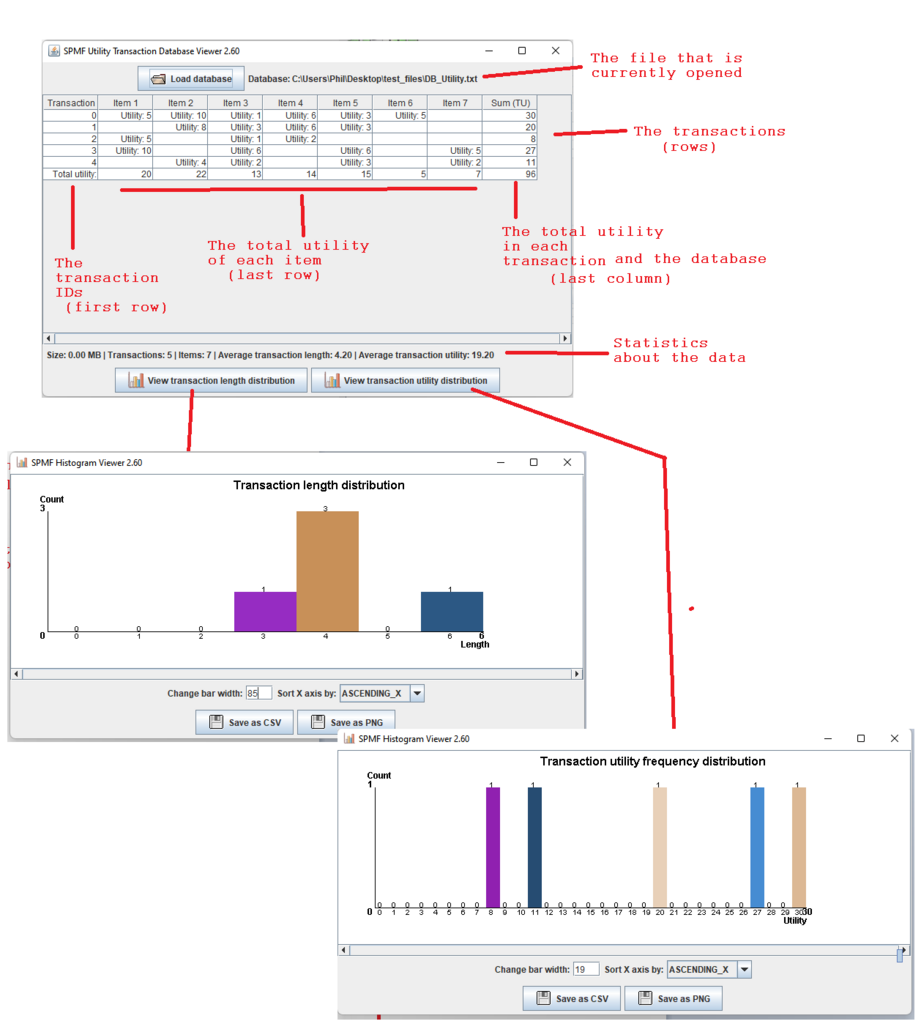
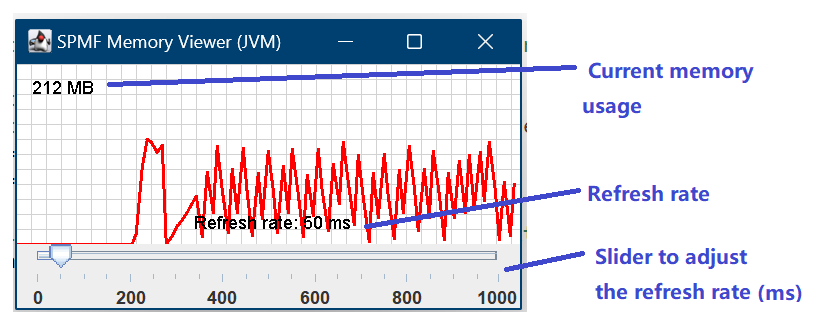
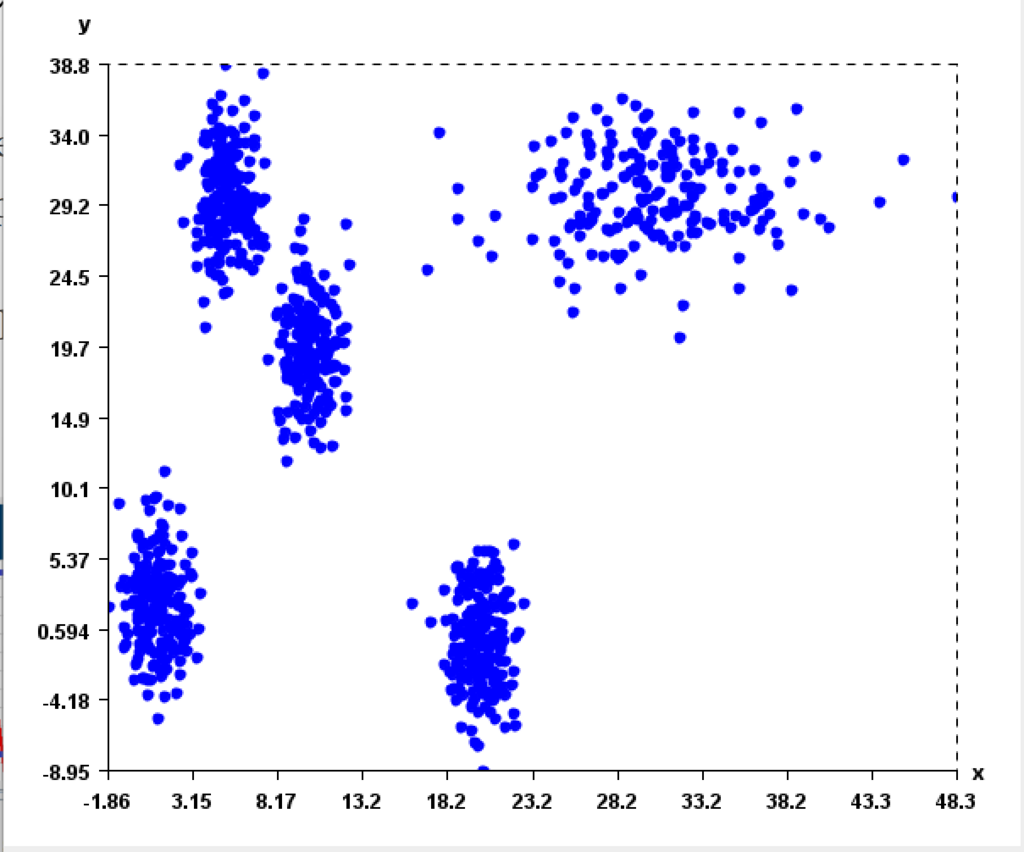
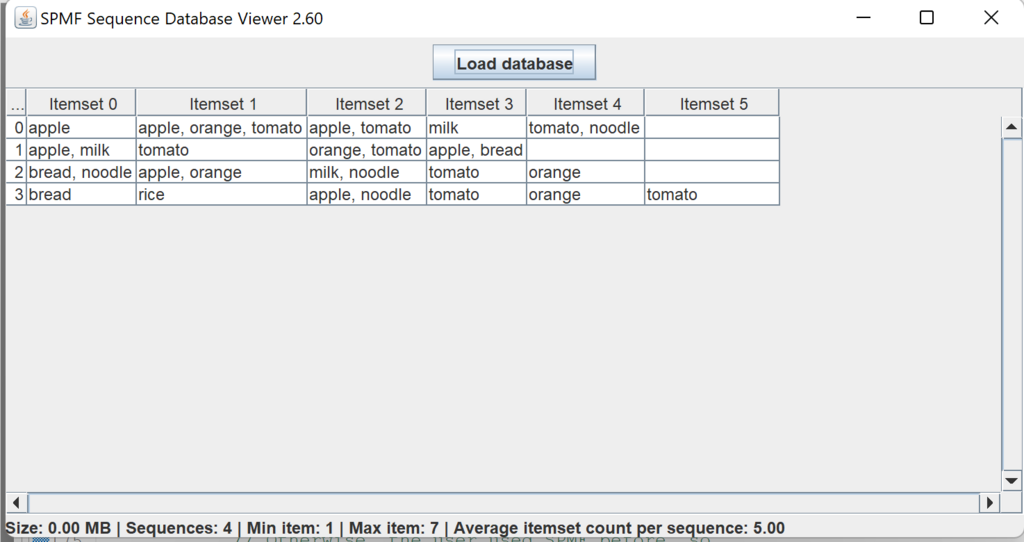
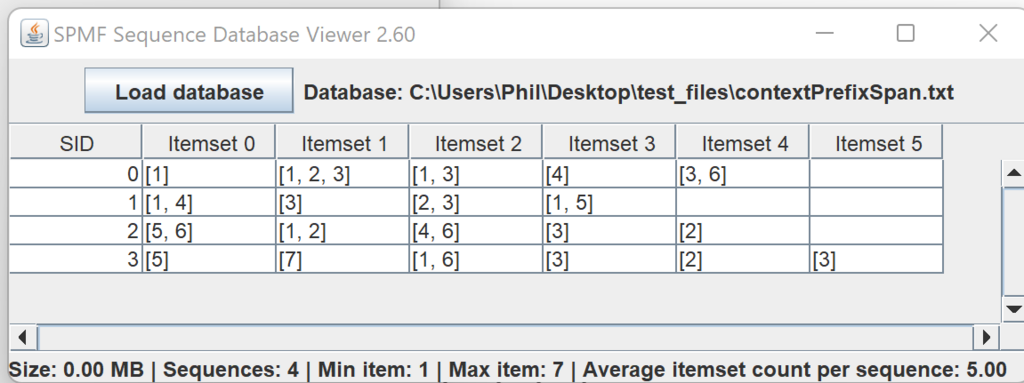
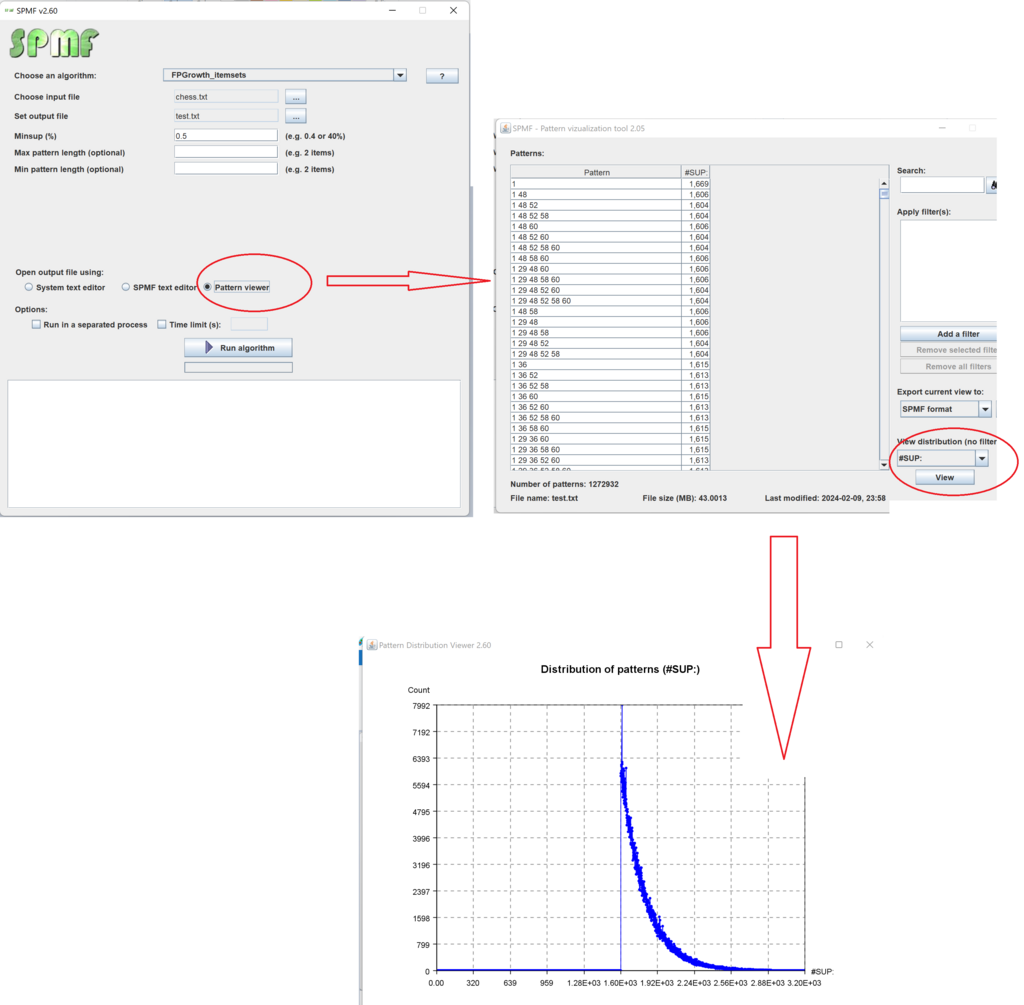
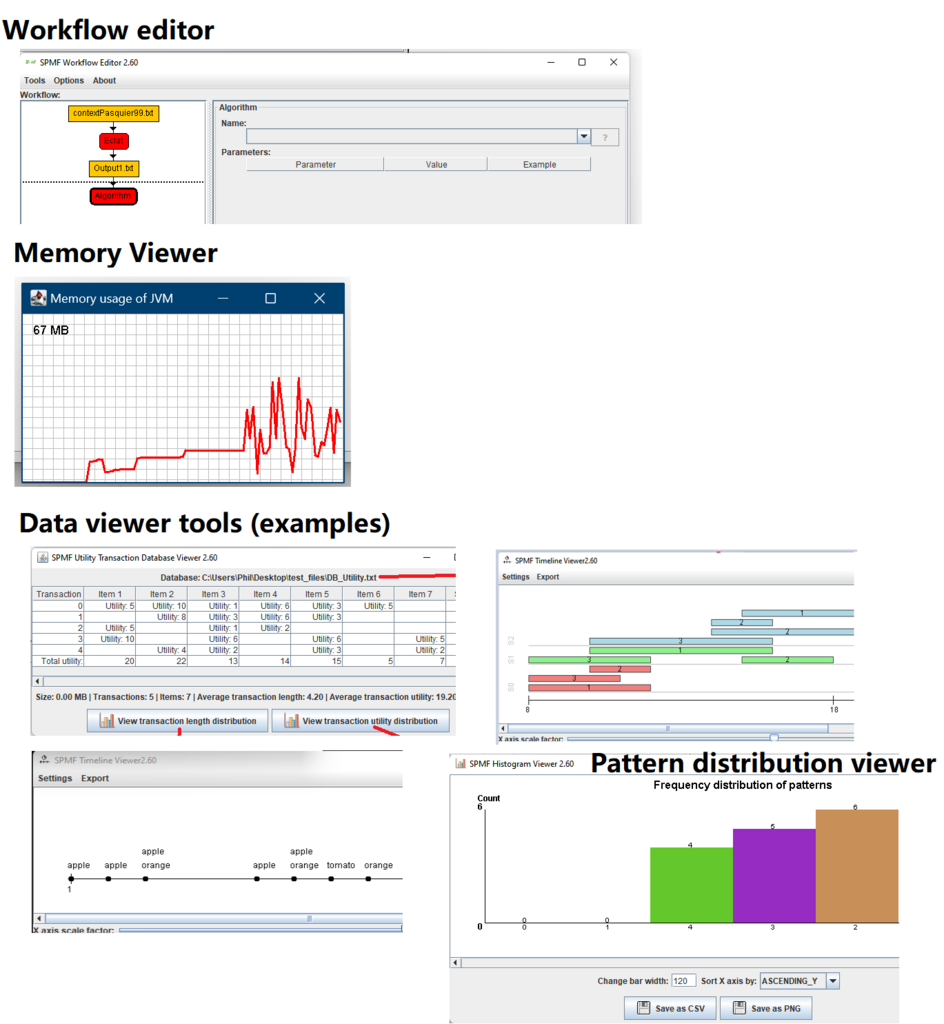
This is a major version as it contains many new things. The full lists of changes can be found on the download page. Some of the main improvements are 18 new algorithms, 21 new tools to visualize different types of data, several improvements to the user interface (some are less visible than others), and also several tools that are added like a workflow editor for running more than one algorithm one after the other, some new tools for data generation and transformation. Here is a picture of a few new windows in the graphical user interface among several:
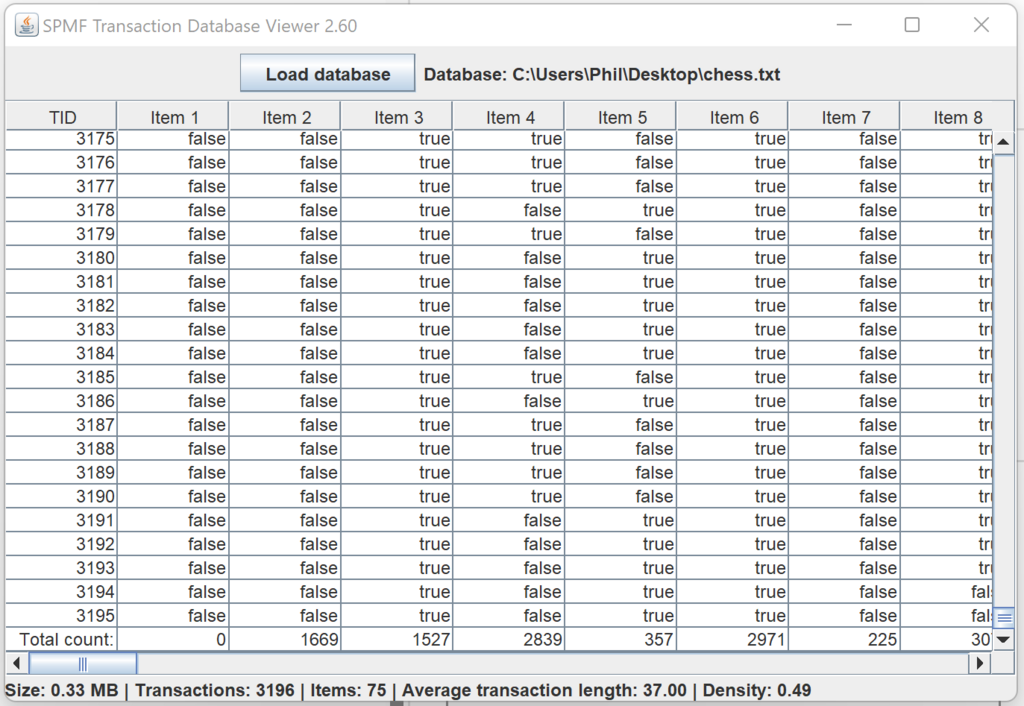
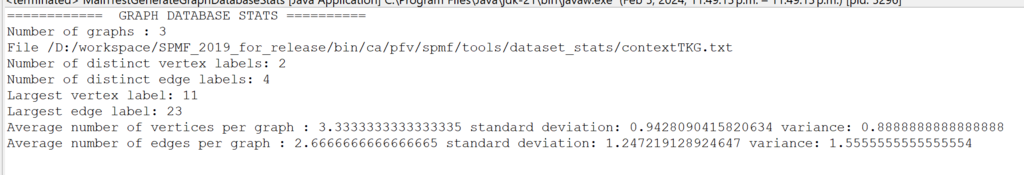
Besides, for developers of algorithms, a collection of new data structures optimized for primitive types (int, double, etc.) are provided in the package ca.pfv.spmf.datastructures.collections, which can replace several standard Java data structures to speed up algorithms or reduce the memory usage. Here is a screenshot of some of those data structures:

I have also fixed several bugs in the software (thanks to all users who reported them). It is possible that some bugs remain, especially because there is a lot of new code. If you find any problems, please let me know at philfv AT qq DOT com. Also, you can let me know about your suggestions for improvements, if you have some ideas. 🙂 If you also want to contribute code to SPMF, please contact with me (for example, if you want that I integrate your algorithm in the software.
Thanks again to all users of SPMF and the contributors, who support this project and make it better.